7O3O
 
 | | Structure of haloalkane dehalogenase mutant DhaA80(T148L, G171Q, A172V, C176F) from Rhodococcus rhodochrous with ionic liquid | | Descriptor: | CHLORIDE ION, ETHANOLAMINE, Haloalkane dehalogenase | | Authors: | Shaposhnikova, A, Prudnikova, T, Kuta Smatanova, I. | | Deposit date: | 2021-04-02 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Stabilization of Haloalkane Dehalogenase Structure by Interfacial Interaction with Ionic Liquids
Crystals, 11, 2021
|
|
7O8B
 
 | |
6PUX
 
 | |
6T6H
 
 | | Apo structure of the Bottromycin epimerase BotH | | Descriptor: | BotH, SODIUM ION, SULFATE ION | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2019-10-18 | | Release date: | 2020-07-15 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | The bottromycin epimerase BotH defines a group of atypical alpha / beta-hydrolase-fold enzymes.
Nat.Chem.Biol., 16, 2020
|
|
6T6Z
 
 | |
6T6Y
 
 | |
4OSE
 
 | |
6QKU
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Tyr219Phe - Chloroacetate soaked 2hr | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, GLYCOLIC ACID, ... | | Authors: | Mehrabi, P, Kim, T.H, Prosser, R.S, Pai, E.F. | | Deposit date: | 2019-01-30 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | Substrate-Based Allosteric Regulation of a Homodimeric Enzyme.
J.Am.Chem.Soc., 141, 2019
|
|
6QHZ
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 6788 ms | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
6QHT
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 376 ms | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
6QI2
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 13536 ms | | Descriptor: | Fluoroacetate dehalogenase, GLYCOLIC ACID | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
6QKW
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Tyr219Phe - Fluoroacetate soaked 2hr | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, GLYCOLIC ACID, ... | | Authors: | Mehrabi, P, Kim, T.H, Prosser, R.S, Pai, E.F. | | Deposit date: | 2019-01-30 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.512 Å) | | Cite: | Substrate-Based Allosteric Regulation of a Homodimeric Enzyme.
J.Am.Chem.Soc., 141, 2019
|
|
6TY7
 
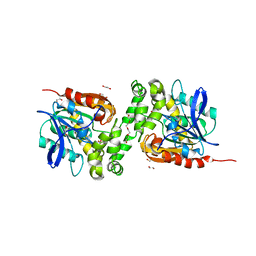 | | Crystal structure of haloalkane dehalogenase variant DhaA115 domain-swapped dimer type-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Markova, K, Chaloupkova, R, Damborsky, J, Marek, M. | | Deposit date: | 2020-01-15 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Computational Enzyme Stabilization Can Affect Folding Energy Landscapes and Lead to Catalytically Enhanced Domain-Swapped Dimers
Acs Catalysis, 11, 2021
|
|
4Q3L
 
 | | Crystal structure of MGS-M2, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | GLYCEROL, MGS-M2 | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
6T70
 
 | |
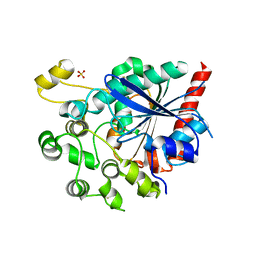
6V7N
 
 | |
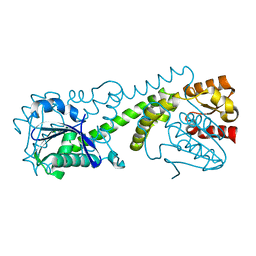
8G48
 
 | |
8G49
 
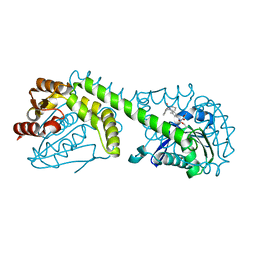 | | FphE, Staphylococcus aureus fluorophosphonate-binding serine hydrolases E, Oxadiazolone compound 3 bound | | Descriptor: | Fluorophosphonate-binding serine hydrolase E, methyl 2-formyl-2-[3-methyl-4-(3-phenoxybenzamido)phenyl]hydrazine-1-carboxylate | | Authors: | Fellner, M, Bakker, A.T, Martin, N.I, Stelt, M. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | FphE, Staphylococcus aureus fluorophosphonate-binding serine hydrolases E, Oxadiazolone compound 3 bound
To be published
|
|
8E18
 
 | | Crystal structure of apo TnmK1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Secreted hydrolase | | Authors: | Liu, Y.-C, Gui, C, Shen, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Intramolecular C-C Bond Formation Links Anthraquinone and Enediyne Scaffolds in Tiancimycin Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
8E19
 
 | | Crystal structure of TnmK1 complexed with TNM H | | Descriptor: | (1R,8S,13S)-8-[(4-hydroxy-9,10-dioxo-9,10-dihydroanthracen-1-yl)amino]-12-methoxy-10-methylbicyclo[7.3.1]trideca-9,11-diene-2,6-diyne-13-carbaldehyde, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SUCCINIC ACID, ... | | Authors: | Liu, Y.-C, Gui, C, Shen, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Intramolecular C-C Bond Formation Links Anthraquinone and Enediyne Scaffolds in Tiancimycin Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
8E5W
 
 | | Crystal structure of dehydroalanine Hip1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PALMITIC ACID, Protease, ... | | Authors: | Goldfarb, N.E, Brooks, C.L, Ostrov, D.A. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.1 angstrom crystal structure of the Mycobacterium tuberculosis serine hydrolase, Hip1, in its anhydro-form (Anhydrohip1).
Biochem.Biophys.Res.Commun., 630, 2022
|
|
8F2L
 
 | |
8HGW
 
 | | Crystal structure of MehpH in complex with MBP | | Descriptor: | 1-BUTANOL, Monoalkyl phthalate hydrolase, PHTHALIC ACID | | Authors: | Zhang, Z.M, Wang, Y.J, Chen, Y.B. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.80001163 Å) | | Cite: | Molecular insights into the catalytic mechanism of plasticizer degradation by a monoalkyl phthalate hydrolase.
Commun Chem, 6, 2023
|
|
8HGV
 
 | |
5ESR
 
 | | Crystal structure of haloalkane dehalogenase (DccA) from Caulobacter crescentus | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Haloalkane dehalogenase, ... | | Authors: | Malashkevich, V.N, Toro, R, Mundorff, E.C, Almo, S.C. | | Deposit date: | 2015-11-17 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.476 Å) | | Cite: | Biochemical characterization of two haloalkane dehalogenases: DccA from Caulobacter crescentus and DsaA from Saccharomonospora azurea.
Protein Sci., 25, 2016
|
|
