7PGI
 
 | | NaVAb1p (bicelles) | | Descriptor: | ACETATE ION, Ion transport protein, MAGNESIUM ION, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.638 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGH
 
 | | NaVAe1/Sp1CTDp (DDM) | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DODECAETHYLENE GLYCOL, DODECANE, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-29 | | Method: | X-RAY DIFFRACTION (4.194 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGG
 
 | | NaVAb1p detergent (DM) | | Descriptor: | 2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}ethyl heptadecanoate, Ion transport protein | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGF
 
 | |
7PGB
 
 | | NaV_Ae1/Sp1CTD_pore-SAT09 complex | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-NITROBENZOIC ACID, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PG8
 
 | | NaV_Ae1/Sp1CTD_pore-ANT05 complex | | Descriptor: | ANT05 H12 fab fragment, heavy chain, light chain, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OR1
 
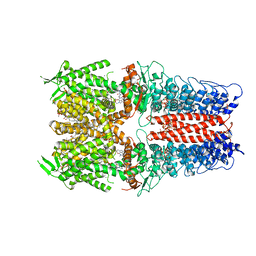 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60, conformation 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grieben, M, Pike, A.C.W, Saward, B.G, Wang, D, Mukhopadhyay, S.M.M, Moreira, T, Chalk, R, MacLean, E.M, Marsden, B.D, Burgess-Brown, N.A, Bountra, C, Schofield, C.J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60
TO BE PUBLISHED
|
|
7OR0
 
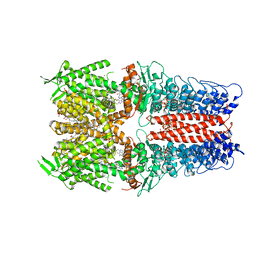 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60, conformation 2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grieben, M, Pike, A.C.W, Saward, B.G, Wang, D, Mukhopadhyay, S.M.M, Moreira, T, Chalk, R, MacLean, E.M, Marsden, B.D, Burgess-Brown, N.A, Bountra, C, Schofield, C.J, Carpenter, E.P. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60
To Be Published
|
|
7O4H
 
 | |
7NP4
 
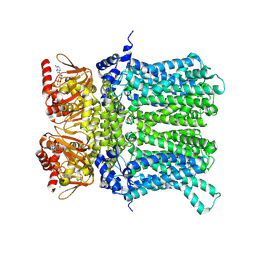 | | cAMP-bound rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Giese, H, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
7NP3
 
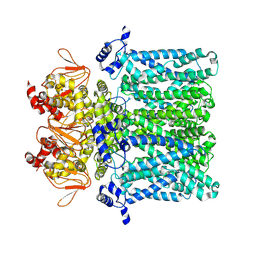 | | cAMP-free rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Giese, H.M, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
7NMN
 
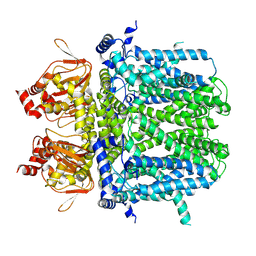 | | Rabbit HCN4 stabilised in amphipol A8-35 | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Rabbit HCN4 | | Authors: | Chaves-Sanjuan, A. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
7N17
 
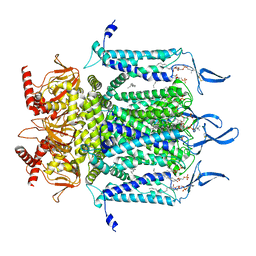 | | Structure of TAX-4_R421W apo open state | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
7N16
 
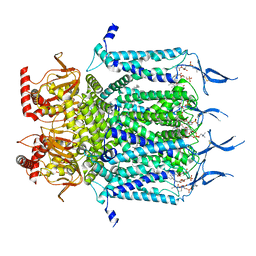 | | Structure of TAX-4_R421W apo closed state | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, SODIUM ION | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
7N15
 
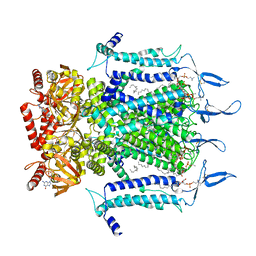 | | Structure of TAX-4_R421W w/cGMP open state | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CYCLIC GUANOSINE MONOPHOSPHATE, ... | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
7N0N
 
 | |
7N0M
 
 | | Inactivated state of 2-APB-bound wildtype rat TRPV2 in nanodiscs | | Descriptor: | 2-aminoethyl diphenylborinate, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Pumroy, R.A, Protopopova, A.D, Gallo, P.N, Moiseenkova-Bell, V.Y. | | Deposit date: | 2021-05-25 | | Release date: | 2022-05-04 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into TRPV2 activation by small molecules.
Nat Commun, 13, 2022
|
|
7MZE
 
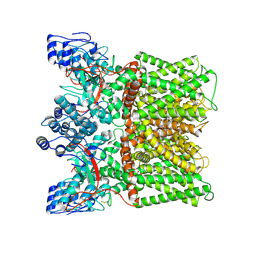 | | Cryo-EM structure of minimal TRPV1 with 2 bound RTX in opposite pockets | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
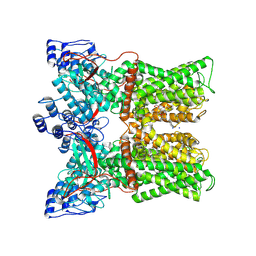
7MZD
 
 | |
7MZC
 
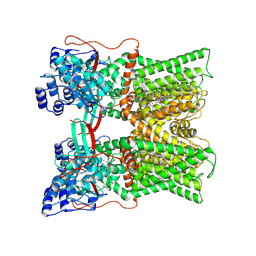 | |
7MZB
 
 | | Cryo-EM structure of minimal TRPV1 with 3 bound RTX and 1 perturbed PI | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZA
 
 | | Cryo-EM structure of minimal TRPV1 with 2 bound RTX in adjacent pockets | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZ9
 
 | | Cryo-EM structure of minimal TRPV1 with 1 partially bound RTX | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZ7
 
 | |
7MZ6
 
 | | Cryo-EM structure of minimal TRPV1 with 1 perturbed PI | | Descriptor: | (10R,13S)-16-amino-13-hydroxy-7,13-dioxo-8,12,14-trioxa-13lambda~5~-phosphahexadecan-10-yl hexadecanoate, (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
