7RSV
 
 | | Structure of the VPS34 kinase domain with compound 5 | | Descriptor: | (5aS,8aR,9S)-2-[(3R)-3-methylmorpholin-4-yl]-5,5a,6,7,8,8a-hexahydro-4H-cyclopenta[e]pyrazolo[1,5-a]pyrazin-4-one, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3, ... | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7RSP
 
 | | Structure of the VPS34 kinase domain with compound 14 | | Descriptor: | (7R,8R)-2-[(3R)-3-methylmorpholin-4-yl]-7-(propan-2-yl)-6,7-dihydropyrazolo[1,5-a]pyrazin-4(5H)-one, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3 | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7PG5
 
 | | Crystal Structure of PI3Kalpha | | Descriptor: | GLYCEROL, PHOSPHATE ION, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gong, G, Pinotsis, N, Williams, R.L, Vanhaesebroeck, B. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.20029068 Å) | | Cite: | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
7PG6
 
 | | Crystal Structure of PI3Kalpha in complex with the inhibitor NVP-BYL719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Gong, G, Pinotsis, N, Williams, R.L, Vanhaesebroeck, B. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49943733 Å) | | Cite: | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
7POT
 
 | | PI3 kinase delta in complex with N-[5-(3,6-dihydro-2H-pyran-4-yl)-2-methoxypyridin-3-yl]benzenesulfonamide | | Descriptor: | N-[5-(3,6-dihydro-2H-pyran-4-yl)-2-methoxy-pyridin-3-yl]benzenesulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Rowland, P, Convery, M. | | Deposit date: | 2021-09-09 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Discovery of GSK251: A Highly Potent, Highly Selective, Orally Bioavailable Inhibitor of PI3K delta with a Novel Binding Mode.
J.Med.Chem., 64, 2021
|
|
7POR
 
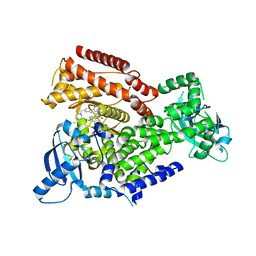 | |
7POP
 
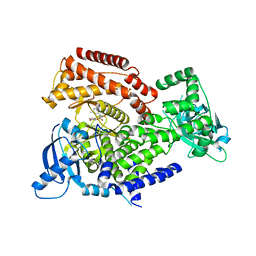 | | PI3 kinase delta in complex with 5-[3,6-dihydro-2H-pyran-4-yl]-2-methoxy-N-[2-methylpyridin-4-yl]pyridine-3-sulfonamide | | Descriptor: | 5-[3,6-dihydro-2H-pyran-4-yl]-2-methoxy-N-[2-methylpyridin-4-yl]pyridine-3-sulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Rowland, P, Convery, M. | | Deposit date: | 2021-09-09 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Discovery of GSK251: A Highly Potent, Highly Selective, Orally Bioavailable Inhibitor of PI3K delta with a Novel Binding Mode.
J.Med.Chem., 64, 2021
|
|
7POS
 
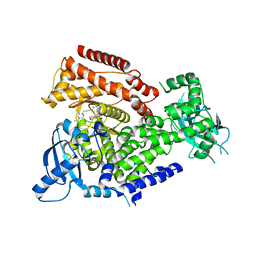 | | PI3 kinase delta in complex with 5-(3,6-dihydro-2H-pyran-4-yl)-2-methoxy-N-(5-{3-[4-(propan-2-yl)piperazin-1-yl]prop-1-yn-1-yl}pyridin-3-yl)pyridine-3-sulfonamide | | Descriptor: | 5-(3,6-dihydro-2~{H}-pyran-4-yl)-2-methoxy-~{N}-[5-[3-(4-propan-2-ylpiperazin-1-yl)prop-1-ynyl]pyridin-3-yl]pyridine-3-sulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Rowland, P, Convery, M. | | Deposit date: | 2021-09-09 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Discovery of GSK251: A Highly Potent, Highly Selective, Orally Bioavailable Inhibitor of PI3K delta with a Novel Binding Mode.
J.Med.Chem., 64, 2021
|
|
7PQH
 
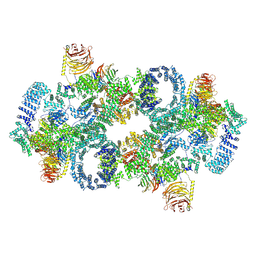 | | Cryo-EM structure of Saccharomyces cerevisiae TOROID (TORC1 Organized in Inhibited Domains). | | Descriptor: | Serine/threonine-protein kinase TOR2, Target of rapamycin complex 1 subunit KOG1,Target of rapamycin complex 1 subunit Kog1, Target of rapamycin complex subunit LST8 | | Authors: | Felix, J, Prouteau, M, Bourgoint, C, Bonadei, L, Desfosses, A, Gabus, C, Sadian, Y, Savvides, S.N, Gutsche, I, Loewith, R. | | Deposit date: | 2021-09-17 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | EGOC inhibits TOROID polymerization by structurally activating TORC1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7SGL
 
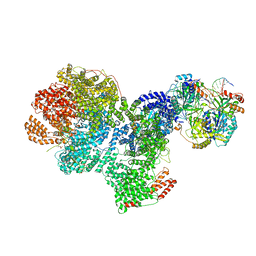 | | DNA-PK complex of DNA end processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, Hairpin_1, ... | | Authors: | Liu, L, Li, J, Chen, X, Yang, W, Gellert, M. | | Deposit date: | 2021-10-06 | | Release date: | 2022-01-12 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
7PW4
 
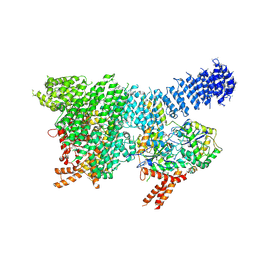 | | Human SMG1-8-9 kinase complex bound to a SMG1 inhibitor | | Descriptor: | 1-[4-[4-[2-[[4-chloranyl-3-(diethylsulfamoyl)phenyl]amino]pyrimidin-4-yl]pyridin-2-yl]phenyl]-3-methyl-urea, ADENOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Langer, L.M, Conti, E. | | Deposit date: | 2021-10-06 | | Release date: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Cryo-EM reconstructions of inhibitor-bound SMG1 kinase reveal an autoinhibitory state dependent on SMG8.
Elife, 10, 2021
|
|
7PW8
 
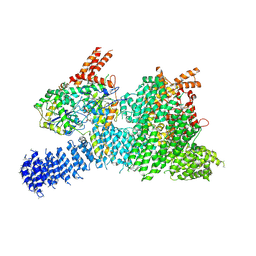 | | Human SMG1-8-9 kinase complex bound to AMPPNP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Langer, L.M, Conti, E. | | Deposit date: | 2021-10-06 | | Release date: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Cryo-EM reconstructions of inhibitor-bound SMG1 kinase reveal an autoinhibitory state dependent on SMG8.
Elife, 10, 2021
|
|
7PW6
 
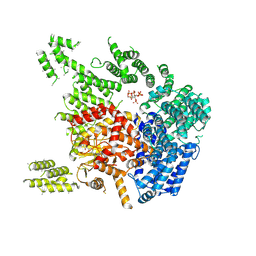 | | Human SMG1-8-9 kinase complex bound to a SMG1 inhibitor - SMG1 body | | Descriptor: | 1-[4-[4-[2-[[4-chloranyl-3-(diethylsulfamoyl)phenyl]amino]pyrimidin-4-yl]pyridin-2-yl]phenyl]-3-methyl-urea, INOSITOL HEXAKISPHOSPHATE, Serine/threonine-protein kinase SMG1,Serine/threonine-protein kinase SMG1,Serine/threonine-protein kinase SMG1,Serine/threonine-protein kinase SMG1,Serine/threonine-protein kinase SMG1,Serine/threonine-protein kinase SMG1,Serine/threonine-protein kinase SMG1 | | Authors: | Langer, L.M, Conti, E. | | Deposit date: | 2021-10-06 | | Release date: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM reconstructions of inhibitor-bound SMG1 kinase reveal an autoinhibitory state dependent on SMG8.
Elife, 10, 2021
|
|
7PW9
 
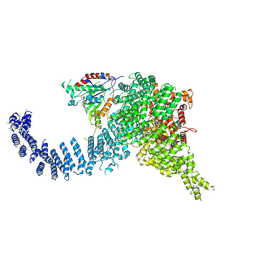 | | Human SMG1-9 kinase complex bound to AMPPNP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Langer, L.M, Conti, E. | | Deposit date: | 2021-10-06 | | Release date: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM reconstructions of inhibitor-bound SMG1 kinase reveal an autoinhibitory state dependent on SMG8.
Elife, 10, 2021
|
|
7PW7
 
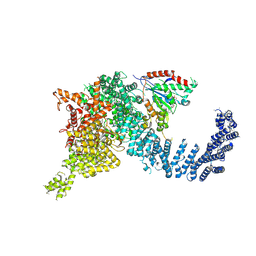 | | Human SMG1-9 kinase complex bound to a SMG1 inhibitor | | Descriptor: | 1-[4-[4-[2-[[4-chloranyl-3-(diethylsulfamoyl)phenyl]amino]pyrimidin-4-yl]pyridin-2-yl]phenyl]-3-methyl-urea, ADENOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Langer, L.M, Conti, E. | | Deposit date: | 2021-10-06 | | Release date: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Cryo-EM reconstructions of inhibitor-bound SMG1 kinase reveal an autoinhibitory state dependent on SMG8.
Elife, 10, 2021
|
|
7PW5
 
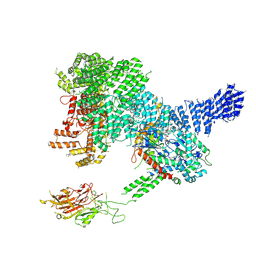 | |
7SIC
 
 | | Human ATM Dimer | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine-protein kinase ATM | | Authors: | Warren, C, Pavletich, N.P. | | Deposit date: | 2021-10-13 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structure of the human ATM kinase and mechanism of Nbs1 binding.
Elife, 11, 2022
|
|
7SID
 
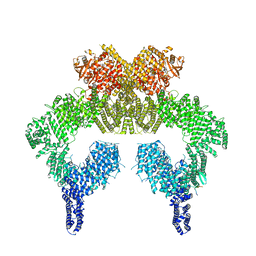 | | Human ATM Dimer Bound to Nbs1 | | Descriptor: | MAGNESIUM ION, Nibrin, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Warren, C, Pavletich, N.P. | | Deposit date: | 2021-10-13 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structure of the human ATM kinase and mechanism of Nbs1 binding.
Elife, 11, 2022
|
|
7VVY
 
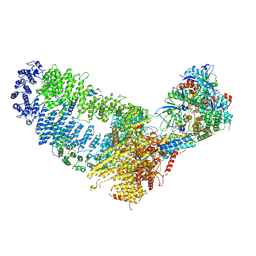 | | TRA module of NuA4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Chen, Z, Qu, K. | | Deposit date: | 2021-11-09 | | Release date: | 2022-08-10 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the NuA4 acetyltransferase complex bound to the nucleosome.
Nature, 610, 2022
|
|
7VVZ
 
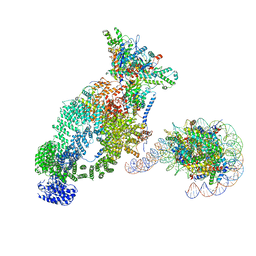 | | NuA4 bound to the nucleosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Qu, K, Chen, Z. | | Deposit date: | 2021-11-09 | | Release date: | 2022-08-10 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structure of the NuA4 acetyltransferase complex bound to the nucleosome.
Nature, 610, 2022
|
|
7SUD
 
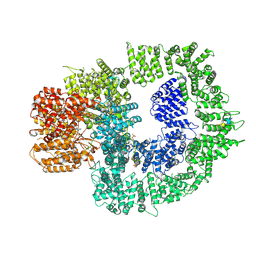 | | CryoEM structure of DNA-PK complex VIII | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, MAGNESIUM ION, ... | | Authors: | Chen, X, Liu, L, Gellert, M, Yang, W. | | Deposit date: | 2021-11-16 | | Release date: | 2022-01-12 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
7SU3
 
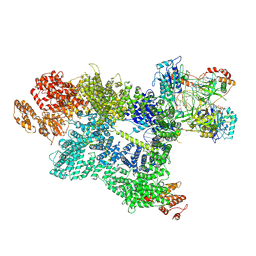 | | CryoEM structure of DNA-PK complex VII | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), ... | | Authors: | Chen, X, Liu, L, Gellert, M, Yang, W. | | Deposit date: | 2021-11-16 | | Release date: | 2022-01-12 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
7R26
 
 | | PI3K delta in complex with SD5 | | Descriptor: | 5-[2,6-di(morpholin-4-yl)pyrimidin-4-yl]-4-(trifluoromethyl)pyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Gutmann, S, Rummel, G, Shrestha, B. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of NVP-CLR457 as an Orally Bioavailable Non-CNS-Penetrant pan-Class IA Phosphoinositol-3-Kinase Inhibitor.
J.Med.Chem., 65, 2022
|
|
7R2B
 
 | | PI3Kdelta in complex with an inhibitor | | Descriptor: | (4~{S})-3-[6-[2-azanyl-4-(trifluoromethyl)pyrimidin-5-yl]-2-morpholin-4-yl-pyrimidin-4-yl]-4-methyl-1,3-oxazolidin-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Gutmann, S, Rummel, G, Shrestha, B. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of NVP-CLR457 as an Orally Bioavailable Non-CNS-Penetrant pan-Class IA Phosphoinositol-3-Kinase Inhibitor.
J.Med.Chem., 65, 2022
|
|
7TYR
 
 | |
