1YJU
 
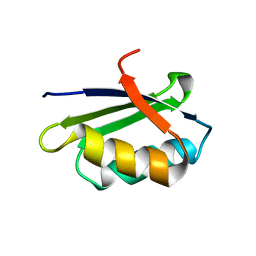 | | Solution structure of the apo form of the sixth soluble domain of Menkes protein | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
1YJT
 
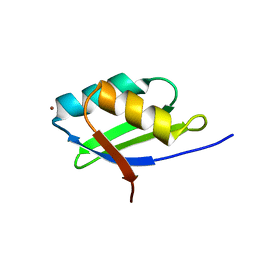 | | Solution structure of the Cu(I) form of the sixth soluble domain A69P mutant of Menkes protein | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S. | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
2AJ1
 
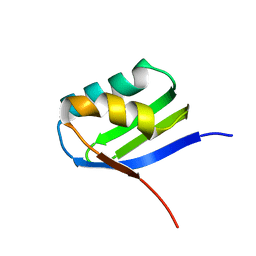 | | Solution structure of apoCadA | | Descriptor: | Probable cadmium-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Su, X.-C, Miras, R, Bal, N, Mintz, E, Catty, P, Shokes, J.E, Scott, R.A. | | Deposit date: | 2005-08-01 | | Release date: | 2006-05-02 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Structural basis for metal binding specificity: the N-terminal cadmium binding domain of the P1-type ATPase CadA
J.Mol.Biol., 356, 2006
|
|
2AJ0
 
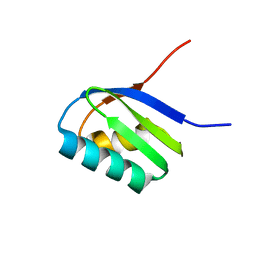 | | Solution structure of apoCadA | | Descriptor: | Probable cadmium-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Su, X.-C, Miras, R, Bal, N, Mintz, E, Catty, P, Shokes, J.E, Scott, R.A. | | Deposit date: | 2005-08-01 | | Release date: | 2006-05-02 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Structural basis for metal binding specificity: the N-terminal cadmium binding domain of the P1-type ATPase CadA
J.Mol.Biol., 356, 2006
|
|
2AW0
 
 | |
1K0V
 
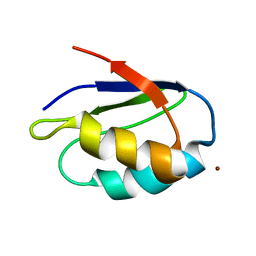 | | Copper trafficking: the solution structure of Bacillus subtilis CopZ | | Descriptor: | COPPER (I) ION, CopZ | | Authors: | Banci, L, Bertini, I, Del Conte, R, Markey, J, Ruiz-Duenas, F.J. | | Deposit date: | 2001-09-21 | | Release date: | 2001-12-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Copper trafficking: the solution structure of Bacillus subtilis CopZ.
Biochemistry, 40, 2001
|
|
1JWW
 
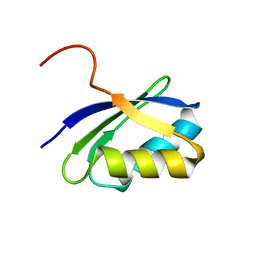 | | NMR characterization of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, D'Onofrio, M, Gonnelli, L, Marhuenda-Egea, F, Ruiz-Duenas, F.J. | | Deposit date: | 2001-09-05 | | Release date: | 2002-04-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis in the apo and Cu(I) loaded states.
J.Mol.Biol., 317, 2002
|
|
1KVJ
 
 | |
1KQK
 
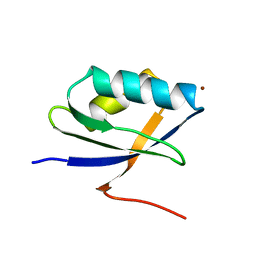 | | Solution Structure of the N-terminal Domain of a Potential Copper-translocating P-type ATPase from Bacillus subtilis in the Cu(I)loaded State | | Descriptor: | COPPER (I) ION, POTENTIAL COPPER-TRANSPORTING ATPASE | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, D'Onofrio, M, Gonnelli, L, Marhuenda-Egea, F.C, Ruiz-Duenas, F.J. | | Deposit date: | 2002-01-07 | | Release date: | 2002-04-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis in the apo and Cu(I) loaded states.
J.Mol.Biol., 317, 2002
|
|
1KVI
 
 | |
1MWY
 
 | | Solution structure of the N-terminal domain of ZntA in the apo-form | | Descriptor: | ZntA | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Finney, L.A, Outten, C.E, O'Halloran, T.V. | | Deposit date: | 2002-10-01 | | Release date: | 2002-11-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A new zinc-protein coordination site in intracellular metal trafficking: solution structure of the apo and Zn(II) forms of ZntA (46-118)
J.Mol.Biol., 323, 2002
|
|
1MWZ
 
 | | Solution structure of the N-terminal domain of ZntA in the Zn(II)-form | | Descriptor: | ZINC ION, ZntA | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Finney, L.A, Outten, C.E, O'Halloran, T.V. | | Deposit date: | 2002-10-01 | | Release date: | 2002-11-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A new zinc-protein coordination site in intracellular metal trafficking: solution structure of the apo and Zn(II) forms of ZntA (46-118)
J.Mol.Biol., 323, 2002
|
|
1P8G
 
 | |
1P6T
 
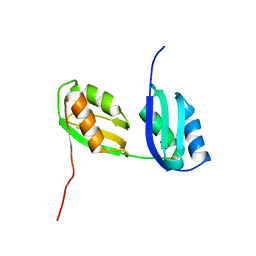 | | Structure characterization of the water soluble region of P-type ATPase CopA from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonnelli, L, Su, X.C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-04-30 | | Release date: | 2003-12-16 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the function of the N-terminal domain of the ATPase CopA from Bacillus subtilis.
J.Biol.Chem., 278, 2003
|
|
1OQ3
 
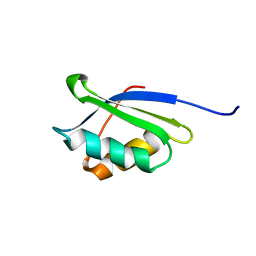 | | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonnelli, L, Su, X.C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-07 | | Release date: | 2003-09-16 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis
J.Mol.Biol., 331, 2003
|
|
2HQI
 
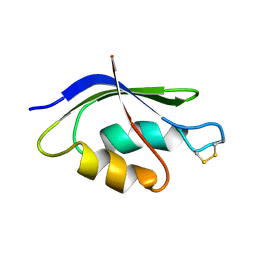 | | NMR SOLUTION STRUCTURE OF THE OXIDIZED FORM OF MERP, 14 STRUCTURES | | Descriptor: | MERCURIC TRANSPORT PROTEIN | | Authors: | Qian, H, Sahlman, L, Eriksson, P.O, Hambreus, C, Edlund, U, Sethson, I. | | Deposit date: | 1998-03-31 | | Release date: | 1998-11-11 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the oxidized form of MerP, a mercuric ion binding protein involved in bacterial mercuric ion resistance.
Biochemistry, 37, 1998
|
|
2K1R
 
 | | The solution NMR structure of the complex between MNK1 and HAH1 mediated by Cu(I) | | Descriptor: | COPPER (II) ION, Copper transport protein ATOX1, Copper-transporting ATPase 1 | | Authors: | Bertini, I, Banci, L.C, Felli, I.C, Pavelkova, A, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-03-14 | | Release date: | 2009-03-31 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the copper(I)-mediated complex between the first soluble domain of the Menkes protein and the metallochaperone HAH1.
To be Published
|
|
2K2P
 
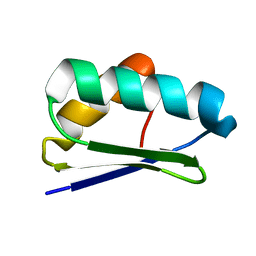 | | Solution NMR structure of protein Atu1203 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium (NESG) target AtT10, Ontario Center for Structural Proteomics target ATC1183 | | Descriptor: | Uncharacterized protein Atu1203 | | Authors: | Lemak, A, Gutmanas, A, Yee, A, Semesi, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-04-08 | | Release date: | 2008-04-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein Atu1203 from Agrobacterium tumefaciens.
To be Published
|
|
2KT2
 
 | | Structure of NmerA, the N-terminal HMA domain of Tn501 Mercuric Reductase | | Descriptor: | Mercuric reductase | | Authors: | Ledwidge, R, Danacea, F, Dotsch, V, Miller, S.M. | | Deposit date: | 2010-01-17 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NmerA of Tn501 mercuric ion reductase: structural modulation of the pKa values of the metal binding cysteine thiols.
Biochemistry, 49, 2010
|
|
2KYZ
 
 | | NMR structure of heavy metal binding protein TM0320 from Thermotoga maritima | | Descriptor: | Heavy metal binding protein | | Authors: | Jaudzems, K, Wahab, A, Serrano, P, Geralt, M, Wuthrich, K, Wilson, I.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-06-09 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of heavy metal binding protein TM0320 from Thermotoga maritima
To be Published
|
|
2KT3
 
 | | Structure of Hg-NmerA, Hg(II) complex of the N-terminal domain of Tn501 Mercuric Reductase | | Descriptor: | MERCURY (II) ION, Mercuric reductase | | Authors: | Miller, S.M, Ledwidge, R, Danacea, F, Dotsch, V. | | Deposit date: | 2010-01-17 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NmerA of Tn501 mercuric ion reductase: structural modulation of the pKa values of the metal binding cysteine thiols.
Biochemistry, 49, 2010
|
|
2L3M
 
 | | Solution structure of the putative copper-ion-binding protein from Bacillus anthracis str. Ames | | Descriptor: | Copper-ion-binding protein | | Authors: | Zhang, Y, Winsor, J, Dubrovska, I, Anderson, W, Radhakrishnan, I, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-16 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | To be published
To be Published
|
|
2LDI
 
 | |
2LQ9
 
 | |
2LQB
 
 | |
