6MWH
 
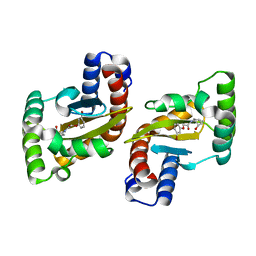 | | LasR LBD:BB0020 complex | | 分子名称: | 2-(3-bromophenoxy)-N-[(1S,2S,3R,5S)-2-hydroxybicyclo[3.1.0]hexan-3-yl]acetamide, Transcriptional regulator LasR | | 著者 | Bassler, B.L, Paczkowski, J.E. | | 登録日 | 2018-10-29 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
6MVN
 
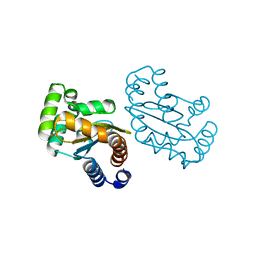 | | LasR LBD L130F:3OC10HSL complex | | 分子名称: | 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]decanamide, Transcriptional regulator LasR | | 著者 | Bassler, B.L, Paczkowski, J.E. | | 登録日 | 2018-10-26 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural determinants driving homoserine lactone ligand selection in thePseudomonas aeruginosaLasR quorum-sensing receptor.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6KJU
 
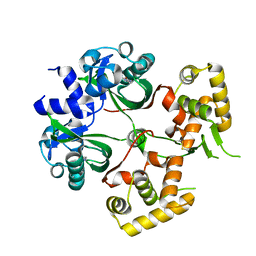 | | Huge conformation shift of Vibrio cholerae VqmA dimer in the absence of target DNA provides insight into DNA-binding mechanisms of LuxR-type receptors | | 分子名称: | 3,5-dimethylpyrazin-2-ol, Helix-turn-helix transcriptional regulator | | 著者 | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Wang, W.W, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | 登録日 | 2019-07-23 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Large conformation shifts of Vibrio cholerae VqmA dimer in the absence of target DNA provide insight into DNA-binding mechanisms of LuxR-type receptors.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
6JQS
 
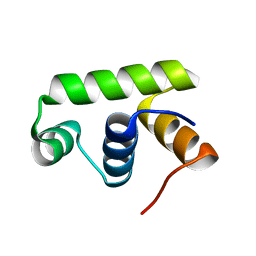 | | Structure of Transcription factor, GerE | | 分子名称: | DNA-binding response regulator | | 著者 | Lee, J.H, Lee, C.W. | | 登録日 | 2019-04-01 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Crystal structure of a transcription factor, GerE (PaGerE), from spore-forming bacterium Paenisporosarcina sp. TG-14.
Biochem.Biophys.Res.Commun., 513, 2019
|
|
6EO3
 
 | |
6EO2
 
 | |
6CC0
 
 | |
6CBQ
 
 | |
5W43
 
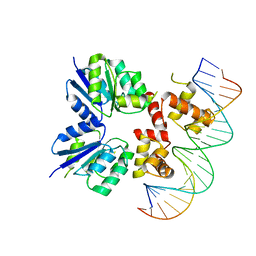 | | Structure of the two-component response regulator RcsB-DNA complex | | 分子名称: | DNA (5'-D(*GP*AP*TP*TP*TP*AP*GP*GP*AP*AP*AP*AP*AP*TP*CP*TP*TP*AP*GP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*CP*TP*AP*AP*GP*AP*TP*TP*TP*TP*TP*CP*CP*TP*AP*AP*AP*TP*C)-3'), Transcriptional regulatory protein RcsB | | 著者 | Filippova, E.V, Warwzak, Z, Pshenychnyi, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-06-09 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structural Basis for DNA Recognition by the Two-Component Response Regulator RcsB.
MBio, 9, 2018
|
|
5VXN
 
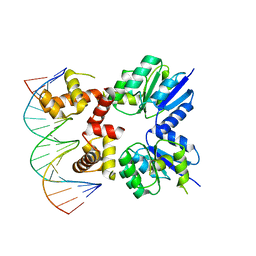 | | Structure of two RcsB dimers bound to two parallel DNAs. | | 分子名称: | DNA (5'-D(*GP*AP*TP*TP*TP*AP*GP*GP*AP*AP*AP*AP*AP*TP*CP*TP*TP*AP*GP*AP*TP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*GP*AP*AP*AP*AP*AP*TP*CP*TP*TP*AP*GP*A)-3'), Transcriptional regulatory protein RcsB | | 著者 | Filippova, E.V, Minasov, G, Pshenychnyi, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-05-23 | | 公開日 | 2018-03-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.375 Å) | | 主引用文献 | Structural Basis for DNA Recognition by the Two-Component Response Regulator RcsB.
MBio, 9, 2018
|
|
5O8Z
 
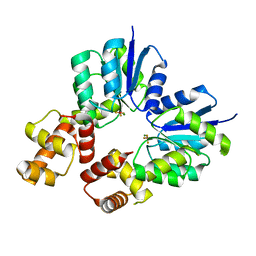 | | Conformational dynamism for DNA interaction in Salmonella typhimurium RcsB response regulator. | | 分子名称: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Transcriptional regulatory protein RcsB | | 著者 | Casino, P, Marina, A, Miguel-Romero, L, Huesa, J. | | 登録日 | 2017-06-14 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Conformational dynamism for DNA interaction in the Salmonella RcsB response regulator.
Nucleic Acids Res., 46, 2018
|
|
5O8Y
 
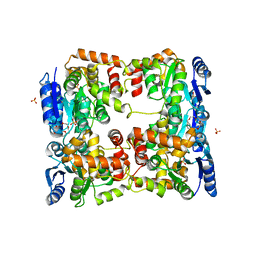 | | Conformational dynamism for DNA interaction in Salmonella typhimurium RcsB response regulator. | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, SULFATE ION, Transcriptional regulatory protein RcsB | | 著者 | Casino, P, Marina, A, Miguel-Romero, L, Huesa, J. | | 登録日 | 2017-06-14 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Conformational dynamism for DNA interaction in the Salmonella RcsB response regulator.
Nucleic Acids Res., 46, 2018
|
|
5F64
 
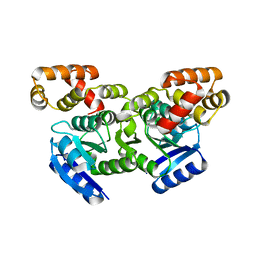 | | Putative positive transcription regulator (sensor EvgS) from Shigella flexneri | | 分子名称: | Positive transcription regulator EvgA | | 著者 | Nocek, B, Osipiuk, J, Mulligan, R, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-12-05 | | 公開日 | 2015-12-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Putative positive transcription regulator (sensor EvgS) from Shigella flexneri.
to be published
|
|
4ZMS
 
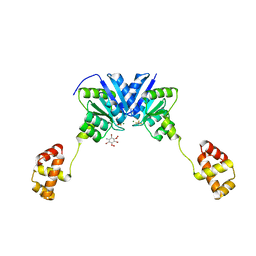 | | Structure of the full-length response regulator spr1814 in complex with a phosphate analogue and B3C | | 分子名称: | 5-amino-2,4,6-tribromobenzene-1,3-diyl dihydroperoxide, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | 著者 | Chi, Y.M, Park, A. | | 登録日 | 2015-05-04 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural characterization of the full-length response regulator spr1814 in complex with a phosphate analogue reveals a novel conformational plasticity of the linker region
Biochem.Biophys.Res.Commun., 473, 2016
|
|
4ZMR
 
 | |
4YN8
 
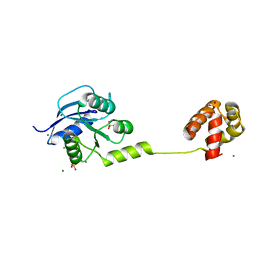 | | Crystal Structure of Response Regulator ChrA in Heme-Sensing Two Component System | | 分子名称: | MAGNESIUM ION, Response regulator ChrA, SULFATE ION | | 著者 | Doi, A, Nakamura, H, Shiro, Y, Sugimoto, H. | | 登録日 | 2015-03-09 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of the response regulator ChrA in the haem-sensing two-component system of Corynebacterium diphtheriae.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4Y17
 
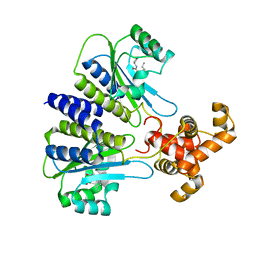 | | SdiA in complex with 3-oxo-C8-homoserine lactone | | 分子名称: | 3-OXO-OCTANOIC ACID (2-OXO-TETRAHYDRO-FURAN-3-YL)-AMIDE, Transcriptional regulator of ftsQAZ gene cluster | | 著者 | Nguyen, N.X, Nguyen, Y, Sperandio, V, Jiang, Y. | | 登録日 | 2015-02-06 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Structural and Mechanistic Roles of Novel Chemical Ligands on the SdiA Quorum-Sensing Transcription Regulator.
Mbio, 6, 2015
|
|
4Y15
 
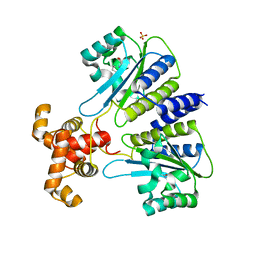 | | SdiA in complex with 3-oxo-C6-homoserine lactone | | 分子名称: | 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide, SULFATE ION, Transcriptional regulator of ftsQAZ gene cluster | | 著者 | Nguyen, N.X, Nguyen, Y, Sperandio, V, Jiang, Y. | | 登録日 | 2015-02-06 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.835 Å) | | 主引用文献 | Structural and Mechanistic Roles of Novel Chemical Ligands on the SdiA Quorum-Sensing Transcription Regulator.
Mbio, 6, 2015
|
|
4Y13
 
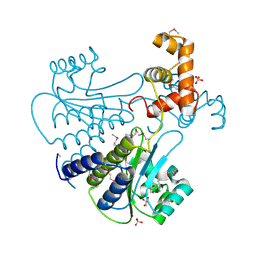 | | SdiA in complex with octanoyl-rac-glycerol | | 分子名称: | (2S)-2,3-dihydroxypropyl octanoate, GLYCEROL, SULFATE ION, ... | | 著者 | Nguyen, N.X, Nguyen, Y, Sperandio, V, Jiang, Y. | | 登録日 | 2015-02-06 | | 公開日 | 2015-04-08 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (3.096 Å) | | 主引用文献 | Structural and Mechanistic Roles of Novel Chemical Ligands on the SdiA Quorum-Sensing Transcription Regulator.
Mbio, 6, 2015
|
|
4LGW
 
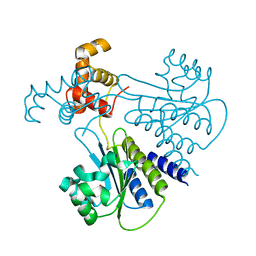 | | Crystal structure of Escherichia coli SdiA in the space group P6522 | | 分子名称: | GLYCEROL, Regulatory protein SdiA | | 著者 | Kim, T, Duong, T, Wu, C.A, Choi, J, Lan, N, Kang, S.W, Lokanath, N.K, Shin, D, Hwang, H.Y, Kim, K.K. | | 登録日 | 2013-06-28 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural insights into the molecular mechanism of Escherichia coli SdiA, a quorum-sensing receptor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LFU
 
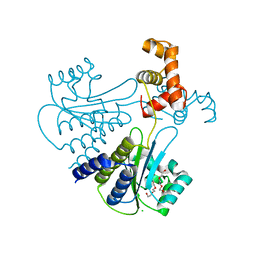 | | Crystal structure of Escherichia coli SdiA in the space group C2 | | 分子名称: | CHLORIDE ION, Regulatory protein SdiA, TETRAETHYLENE GLYCOL | | 著者 | Kim, T, Duong, T, Wu, C.A, Choi, J, Lan, N, Kang, S.W, Lokanath, N.K, Shin, D, Hwang, H.Y, Kim, K.K. | | 登録日 | 2013-06-27 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Structural insights into the molecular mechanism of Escherichia coli SdiA, a quorum-sensing receptor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LDZ
 
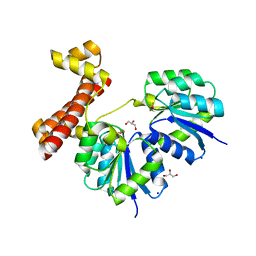 | | Crystal structure of the full-length response regulator DesR in the active state | | 分子名称: | BERYLLIUM TRIFLUORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | 登録日 | 2013-06-25 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation.
MBio, 5, 2014
|
|
4IF4
 
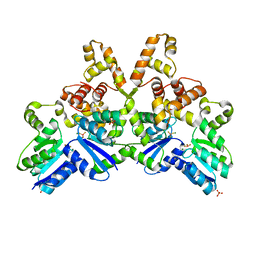 | |
4HYE
 
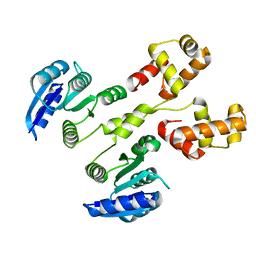 | |
4GVP
 
 | |
