8EB0
 
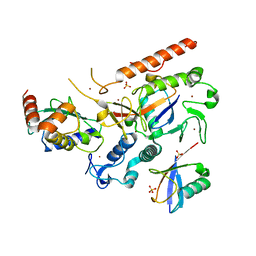 | | RNF216/E2-Ub/Ub transthiolation complex | | Descriptor: | E3 ubiquitin-protein ligase RNF216, SULFATE ION, Ubiquitin, ... | | Authors: | Cotton, T.R, Wang, X.S, Lechtenberg, B.C. | | Deposit date: | 2022-08-30 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | The unifying catalytic mechanism of the RING-between-RING E3 ubiquitin ligase family.
Nat Commun, 14, 2023
|
|
8E2D
 
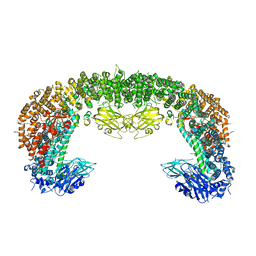 | | Cryo-EM structure of BIRC6 (consensus) | | Descriptor: | Baculoviral IAP repeat-containing protein 6 | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.07 Å) | | Cite: | Structures of BIRC6-client complexes provide a mechanism of SMAC-mediated release of caspases.
Science, 379, 2023
|
|
8E2E
 
 | |
8E2G
 
 | |
8E2I
 
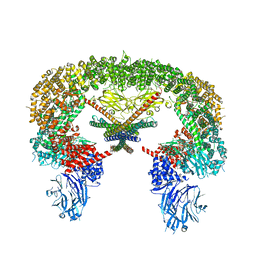 | | Cryo-EM structure of BIRC6/Smac | | Descriptor: | Baculoviral IAP repeat-containing protein 6, Diablo IAP-binding mitochondrial protein | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structures of BIRC6-client complexes provide a mechanism of SMAC-mediated release of caspases.
Science, 379, 2023
|
|
8E2F
 
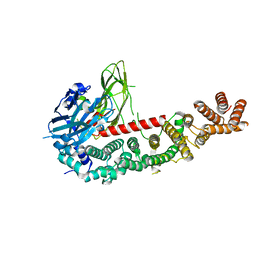 | |
8E2K
 
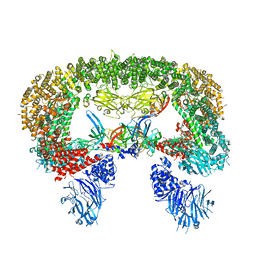 | | Cryo-EM structure of BIRC6/HtrA2-S306A | | Descriptor: | Baculoviral IAP repeat-containing protein 6, Serine protease HTRA2, mitochondrial | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structures of BIRC6-client complexes provide a mechanism of SMAC-mediated release of caspases.
Science, 379, 2023
|
|
8E2H
 
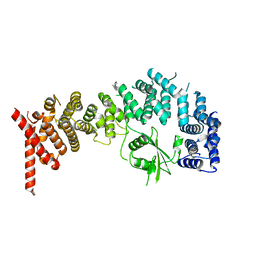 | |
4ORH
 
 | | Crystal structure of RNF8 bound to the UBC13/MMS2 heterodimer | | Descriptor: | E3 ubiquitin-protein ligase RNF8, Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 2, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2014-02-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.802 Å) | | Cite: | Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation.
J.Biol.Chem., 287, 2012
|
|
3A4S
 
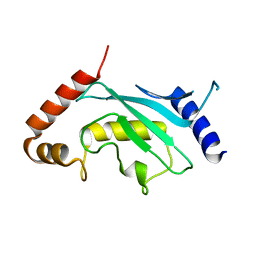 | | The crystal structure of the SLD2:Ubc9 complex | | Descriptor: | NFATC2-interacting protein, SUMO-conjugating enzyme UBC9 | | Authors: | Sekiyama, N, Arita, K, Ikeda, Y, Ariyoshi, M, Tochio, H, Saitoh, H, Shirakawa, M. | | Deposit date: | 2009-07-14 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for regulation of poly-SUMO chain by a SUMO-like domain of Nip45
Proteins, 78, 2009
|
|
4Q5H
 
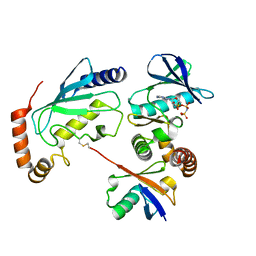 | |
4Q5E
 
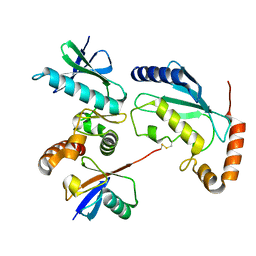 | |
2XWU
 
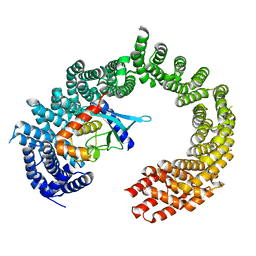 | | CRYSTAL STRUCTURE OF IMPORTIN 13 - UBC9 COMPLEX | | Descriptor: | IMPORTIN13, SUMO-CONJUGATING ENZYME UBC9 | | Authors: | Gruenwald, M, Bono, F. | | Deposit date: | 2010-11-05 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Importin13-Ubc9 Complex: Nuclear Import and Release of a Key Regulator of Sumoylation.
Embo J., 30, 2011
|
|
2YHO
 
 | | The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor | | Descriptor: | ACETATE ION, E3 UBIQUITIN-PROTEIN LIGASE MYLIP, UBIQUITIN-CONJUGATING ENZYME E2 D1, ... | | Authors: | Fairall, L, Goult, B.T, Millard, C.J, Tontonoz, P, Schwabe, J.W.R. | | Deposit date: | 2011-05-04 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Idol-Ube2D Complex Mediates Sterol-Dependent Degradation of the Ldl Receptor.
Genes Dev., 25, 2011
|
|
8GCB
 
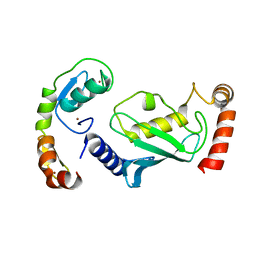 | | Structure of RNF125 in complex with a UbcH5b~Ub conjugate | | Descriptor: | E3 ubiquitin-protein ligase RNF125, Ubiquitin-conjugating enzyme E2 D2, ZINC ION | | Authors: | Middleton, A.J, Day, C.L, Fokkens, T.J. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Zinc finger 1 of the RING E3 ligase, RNF125, interacts with the E2 to enhance ubiquitylation.
Structure, 31, 2023
|
|
8GBQ
 
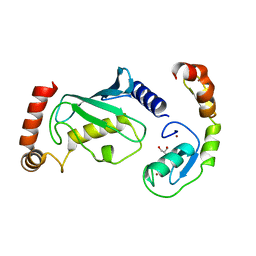 | | Structure of RNF125 in complex with UbcH5b | | Descriptor: | E3 ubiquitin-protein ligase RNF125, GLYCEROL, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Middleton, A.J, Day, C.L, Fokkens, T.J. | | Deposit date: | 2023-02-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Zinc finger 1 of the RING E3 ligase, RNF125, interacts with the E2 to enhance ubiquitylation.
Structure, 31, 2023
|
|
4QPL
 
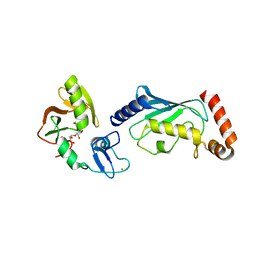 | | Crystal structure of RNF146(RING-WWE)/UbcH5a/iso-ADPr complex | | Descriptor: | 2'-O-(5-O-phosphono-alpha-D-ribofuranosyl)adenosine 5'-(dihydrogen phosphate), E3 ubiquitin-protein ligase RNF146, Ubiquitin-conjugating enzyme E2 D1, ... | | Authors: | Wang, Z, DaRosa, P.A, Klevit, R.E, Xu, W. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal.
Nature, 517, 2015
|
|
4R62
 
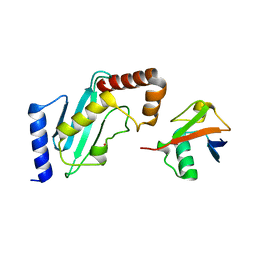 | | Structure of Rad6~Ub | | Descriptor: | ACETATE ION, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 2 | | Authors: | Kumar, P, Wolberger, C. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Role of a non-canonical surface of Rad6 in ubiquitin conjugating activity.
Nucleic Acids Res., 43, 2015
|
|
4TKP
 
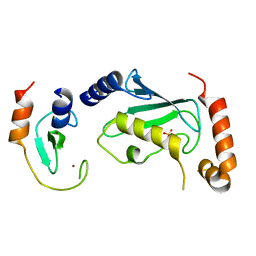 | | Complex of Ubc13 with the RING domain of the TRIM5alpha retroviral restriction factor | | Descriptor: | SULFATE ION, Tripartite motif-containing protein 5, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Johnson, R, Taylor, A.B, Hart, P.J, Ivanov, D.N. | | Deposit date: | 2014-05-27 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | RING Dimerization Links Higher-Order Assembly of TRIM5 alpha to Synthesis of K63-Linked Polyubiquitin.
Cell Rep, 12, 2015
|
|
4S3O
 
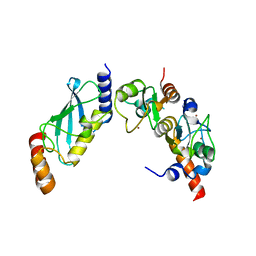 | | PCGF5-RING1B-UbcH5c complex | | Descriptor: | E3 ubiquitin-protein ligase RING2, Polycomb group RING finger protein 5, Ubiquitin-conjugating enzyme E2 D3, ... | | Authors: | Taherbhoy, A.M, Cochran, A.G. | | Deposit date: | 2015-03-23 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | BMI1-RING1B is an autoinhibited RING E3 ubiquitin ligase.
Nat Commun, 6, 2015
|
|
4P5O
 
 | |
3A33
 
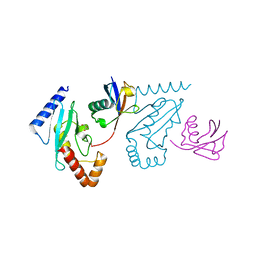 | | UbcH5b~Ubiquitin Conjugate | | Descriptor: | GLYCEROL, Ubiquitin, Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Sakata, E, Satoh, T, Yamamoto, S, Yamaguchi, Y, Yagi-Utsumi, M, Kurimoto, E, Wakatsuki, S, Kato, K. | | Deposit date: | 2009-06-08 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of UbcH5b~Ubiquitin Intermediate: Insight into the Formation of the Self-Assembled E2~Ub Conjugates
Structure, 18, 2010
|
|
2Y9P
 
 | | Pex4p-Pex22p mutant II structure | | Descriptor: | PEROXISOME ASSEMBLY PROTEIN 22, UBIQUITIN-CONJUGATING ENZYME E2-21 KDA | | Authors: | Williams, C, van den Berg, M, Panjikar, S, Distel, B, Wilmanns, M. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Pex4P-Pex22P Structure
To be Published
|
|
4BVU
 
 | | Structure of Shigella effector OspG in complex with host UbcH5c- Ubiquitin conjugate | | Descriptor: | PROTEIN KINASE OSPG, UBIQUITIN, UBIQUITIN-CONJUGATING ENZYME E2 D3 | | Authors: | Pruneda, J.N, LeTrong, I, Stenkamp, R.E, Klevit, R.E, Brzovic, P.S. | | Deposit date: | 2013-06-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | E2~Ub Conjugates Regulate the Kinase Activity of Shigella Effector Ospg During Pathogenesis.
Embo J., 33, 2014
|
|
4CCG
 
 | | Structure of an E2-E3 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Hodson, C, Purkiss, A, Walden, H. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Human Fancl Ring-Ube2T Complex Reveals Determinants of Cognate E3-E2 Selection.
Structure, 22, 2014
|
|
