8WH8
 
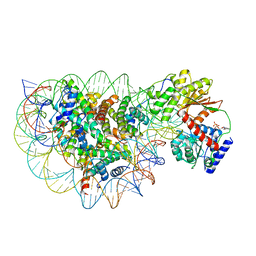 | | Structure of DDM1-nucleosome complex in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, DNA (antisense strand), ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH9
 
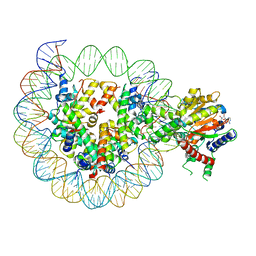 | | Structure of DDM1-nucleosome complex in ADP-BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WHB
 
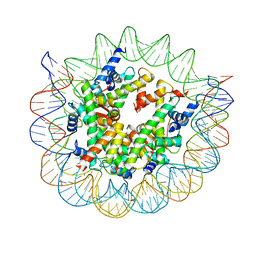 | | Structure of nucleosome core particle of Arabidopsis thaliana | | Descriptor: | DNA (antisense strand), DNA (sense strand), Histone H2A.6, ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-23 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WHA
 
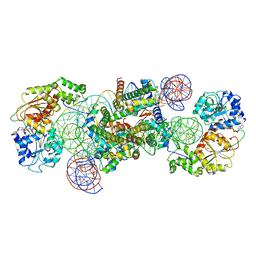 | | Structure of DDM1-nucleosome complex in the ADP-BeFx state with DDM1 bound to SHL2 and SHL-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH5
 
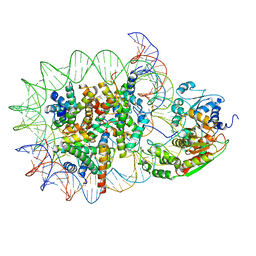 | | Structure of DDM1-nucleosome complex in the apo state | | Descriptor: | ATP-dependent DNA helicase DDM1, DNA (antisense strand), DNA (sense strand), ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8YTI
 
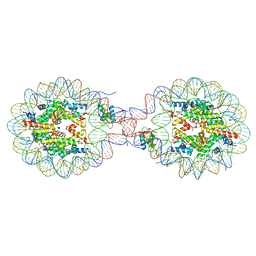 | | Crystal Structure of Nucleosome-H1x Linker Histone Assembly (sticky-169a DNA fragment) | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (169-MER), ... | | Authors: | Adhireksan, Z, Qiuye, B, Padavattan, S, Davey, C.A. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Linker Histones Associate Heterogeneously with Nucleosomes in the Condensed State
To Be Published
|
|
8PP6
 
 | |
8XBT
 
 | | The cryo-EM structure of the octameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JNE
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome without the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JND
 
 | | The cryo-EM structure of the nonameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JNF
 
 | | The cryo-EM structure of the RAD51 filament bound to the nucleosome | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.91 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBU
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBW
 
 | | The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*CP*CP*GP*CP*TP*TP*AP*AP*AP*CP*GP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*CP*GP*TP*TP*TP*AP*AP*GP*CP*GP*GP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8Q3X
 
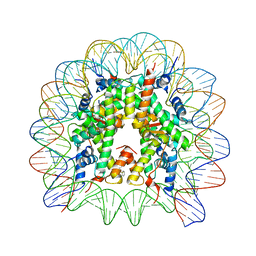 | | Structure of Nucleosome Core with a Bound Metallopeptide Conjugate (Kaposi Sarcoma Associated Herpesvirus LANA Peptide-Au[I] Compound) | | Descriptor: | 4-diphenylphosphanylbenzoic acid, DNA (145-MER), GOLD ION, ... | | Authors: | De Falco, L, Batchelor, L.K, Dyson, P.J, Davey, C.A. | | Deposit date: | 2023-08-04 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Viral peptide conjugates for metal-warhead delivery to chromatin.
Rsc Adv, 14, 2024
|
|
8Q3E
 
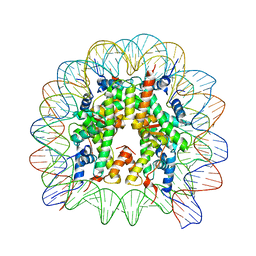 | | High Resolution Structure of Nucleosome Core with Bound Foamy Virus GAG Peptide | | Descriptor: | DNA (145-MER), GLY-GLY-TYR-ASN-LEU-ARG-PRO-ARG-THR-TYR-GLN-PRO-GLN-ARG-TYR-GLY-GLY-GLY, Histone H2A type 1-B/E, ... | | Authors: | De Falco, L, Batchelor, L.K, Dyson, P.J, Davey, C.A. | | Deposit date: | 2023-08-04 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Viral peptide conjugates for metal-warhead delivery to chromatin.
Rsc Adv, 14, 2024
|
|
8Q3M
 
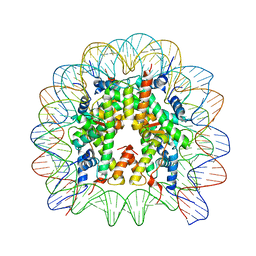 | | Structure of Nucleosome Core with a Bound Kaposi Sarcoma Associated Herpesvirus LANA Peptide Having a Methionine to Ornithine Substitution | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | De Falco, L, Batchelor, L.K, Dyson, P.J, Davey, C.A. | | Deposit date: | 2023-08-04 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Viral peptide conjugates for metal-warhead delivery to chromatin.
Rsc Adv, 14, 2024
|
|
8PP7
 
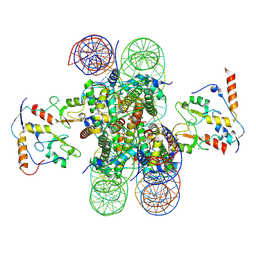 | | human RYBP-PRC1 bound to mononucleosome | | Descriptor: | DNA (215-mer), E3 ubiquitin-protein ligase RING2, Histone H2A, ... | | Authors: | Ciapponi, M, Benda, C, Mueller, J. | | Deposit date: | 2023-07-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis of the histone ubiquitination read-write mechanism of RYBP-PRC1.
Nat.Struct.Mol.Biol., 2024
|
|
8Q36
 
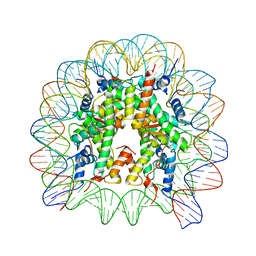 | | Structure of Nucleosome Core with a Bound Metallopeptide Conjugate (Foamy Virus GAG Peptide-Au[I] Compound) | | Descriptor: | DNA (145-MER), GAG structural protein, Histone H2A type 1-B/E, ... | | Authors: | De Falco, L, Batchelor, L.K, Dyson, P.J, Davey, C.A. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Viral peptide conjugates for metal-warhead delivery to chromatin.
Rsc Adv, 14, 2024
|
|
8V7L
 
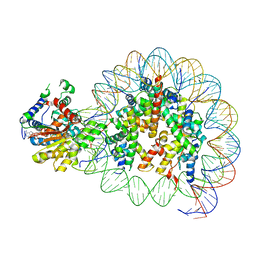 | | Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Histone H2A type 1, Histone H2B, ... | | Authors: | Chio, U.S, Palovcak, E, Armache, J.P, Narlikar, G.J, Cheng, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Nat Commun, 15, 2024
|
|
8V6V
 
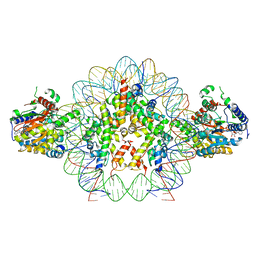 | | Cryo-EM structure of doubly-bound SNF2h-nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Histone H2A type 1, Histone H2B, ... | | Authors: | Chio, U.S, Palovcak, E, Armache, J.P, Narlikar, G.J, Cheng, Y. | | Deposit date: | 2023-12-03 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Nat Commun, 15, 2024
|
|
8V4Y
 
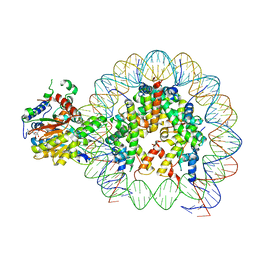 | | Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Histone H2A type 1, ... | | Authors: | Chio, U.S, Palovcak, E, Armache, J.P, Narlikar, G.J, Cheng, Y. | | Deposit date: | 2023-11-29 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Nat Commun, 15, 2024
|
|
8YJM
 
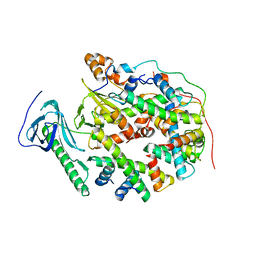 | | Structure of human SPT16 MD-CTD and MCM2 HBD chaperoning a histone H3-H4 tetramer and a single chain H2B-H2A chimera | | Descriptor: | DNA replication licensing factor MCM2, FACT complex subunit SPT16, Histone H2B 1/2/3/4/6,Histone H2A type 1-D, ... | | Authors: | Gan, S.L, Yang, W.S, Xu, R.M. | | Deposit date: | 2024-03-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Structure of a histone hexamer bound by the chaperone domains of SPT16 and MCM2.
Sci China Life Sci, 2024
|
|
8YJF
 
 | |
8QU4
 
 | | NF-YB/C Heterodimer in Complex with a 13-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker in an alternative binding pose | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nuclear transcription factor Y subunit alpha, ... | | Authors: | Arbore, F, Durukan, C, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-10-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
8QU2
 
 | | NF-YB/C Heterodimer in Complex with a 16-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, ... | | Authors: | Durukan, C, Arbore, F, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-10-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
