8FVZ
 
 | | PiPT Y150A | | Descriptor: | CITRATE ANION, PHOSPHATE ION, Phosphate transporter | | Authors: | Gupta, M, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2023-01-20 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Roles of PiPT residues in phosphate binding and transport tested by mutagenesis
To be published
|
|
4PYP
 
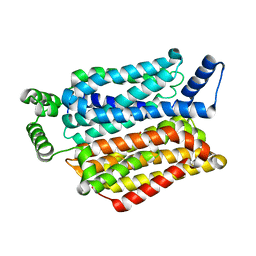 | | Crystal structure of the human glucose transporter GLUT1 | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 1, nonyl beta-D-glucopyranoside | | Authors: | Deng, D, Yan, C.Y, Xu, C, Wu, J.P, Sun, P.C, Hu, M.X, Yan, N. | | Deposit date: | 2014-03-27 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.166 Å) | | Cite: | Crystal structure of the human glucose transporter GLUT1
Nature, 510, 2014
|
|
7AAR
 
 | | sugar/H+ symporter STP10 in inward open conformation | | Descriptor: | CHLORIDE ION, Octyl Glucose Neopentyl Glycol, Sugar transport protein 10, ... | | Authors: | Bavnhoej, L, Paulsen, P.A, Pedersen, B.P. | | Deposit date: | 2020-09-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Molecular mechanism of sugar transport in plants unveiled by structures of glucose/H + symporter STP10.
Nat.Plants, 7, 2021
|
|
7AAQ
 
 | | sugar/H+ symporter STP10 in outward occluded conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, ACETATE ION, ... | | Authors: | Bavnhoej, L, Paulsen, P.A, Pedersen, B.P. | | Deposit date: | 2020-09-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Molecular mechanism of sugar transport in plants unveiled by structures of glucose/H + symporter STP10.
Nat.Plants, 7, 2021
|
|
6GV1
 
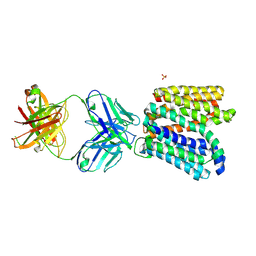 | | Crystal structure of E.coli Multidrug/H+ antiporter MdfA in outward open conformation with bound Fab fragment | | Descriptor: | Fab fragment YN1074 heavy chain, Fab fragment YN1074 light chain, Major Facilitator Superfamily multidrug/H+ antiporter MdfA from E.coli, ... | | Authors: | Nagarathinam, K, Parthier, C, Stubbs, M.T, Tanabe, M. | | Deposit date: | 2018-06-20 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Outward open conformation of a Major Facilitator Superfamily multidrug/H+antiporter provides insights into switching mechanism.
Nat Commun, 9, 2018
|
|
6S4M
 
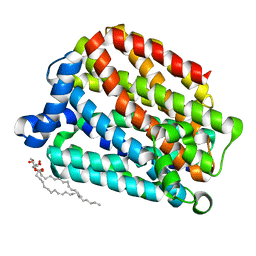 | | Crystal structure of the human organic anion transporter MFSD10 (TETRAN) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRIC ACID, Major facilitator superfamily domain-containing protein 10 | | Authors: | Pascoa, T.C, Pike, A.C.W, Bushell, S.R, Quigley, A, Chu, A, Mukhopadhyay, S.M.M, Shrestha, L, Venkaya, S, Chalk, R, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-28 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human organic anion transporter TETRAN (MFSD10)
To be published
|
|
3O7P
 
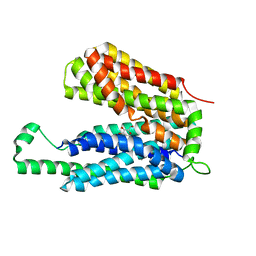 | | Crystal structure of the E.coli Fucose:proton symporter, FucP (N162A) | | Descriptor: | L-fucose-proton symporter, nonyl beta-D-glucopyranoside | | Authors: | Dang, S.Y, Sun, L.F, Wang, J, Yan, N. | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Structure of a fucose transporter in an outward-open conformation
Nature, 467, 2010
|
|
3O7Q
 
 | | Crystal structure of a Major Facilitator Superfamily (MFS) transporter, FucP, in the outward conformation | | Descriptor: | L-fucose-proton symporter, nonyl beta-D-glucopyranoside | | Authors: | Sun, L.F, Dang, S.Y, Wang, J, Yan, N. | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.143 Å) | | Cite: | Structure of a fucose transporter in an outward-open conformation
Nature, 467, 2010
|
|
4QIQ
 
 | | Crystal structure of D-xylose-proton symporter | | Descriptor: | D-xylose-proton symporter, ZINC ION | | Authors: | Wisedchaisri, G, Park, M, Iadanza, M.G, Zheng, H, Gonen, T. | | Deposit date: | 2014-06-01 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Proton-coupled sugar transport in the prototypical major facilitator superfamily protein XylE.
Nat Commun, 5, 2014
|
|
4YB9
 
 | | Crystal structure of the Bovine Fructose transporter GLUT5 in an open inward-facing conformation | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 5 | | Authors: | Verdon, G, Kang, H.J, Iwata, S, Drew, D. | | Deposit date: | 2015-02-18 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanism of the mammalian fructose transporter GLUT5.
Nature, 526, 2015
|
|
4YBQ
 
 | | Rat GLUT5 with Fv in the outward-open form | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 5, antibody Fv fragment heavy chain, ... | | Authors: | Nomura, N, Shimamura, T, Iwata, S. | | Deposit date: | 2015-02-19 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Structure and mechanism of the mammalian fructose transporter GLUT5
Nature, 526, 2015
|
|
6RW3
 
 | | The molecular basis for sugar import in malaria parasites. | | Descriptor: | Hexose transporter 1, alpha-D-glucopyranose, beta-D-glucopyranose | | Authors: | Qureshi, A, Matsuoka, R, Brock, J, Drew, D. | | Deposit date: | 2019-06-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | The molecular basis for sugar import in malaria parasites.
Nature, 578, 2020
|
|
6H7D
 
 | | Crystal Structure of A. thaliana Sugar Transport Protein 10 in complex with glucose in the outward occluded state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, PHOSPHATE ION, ... | | Authors: | Pedersen, B.P, Paulsen, P.A, Custodio, T.F. | | Deposit date: | 2018-07-31 | | Release date: | 2019-02-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the plant symporter STP10 illuminates sugar uptake mechanism in monosaccharide transporter superfamily.
Nat Commun, 10, 2019
|
|
8OMU
 
 | |
7WSM
 
 | | Cryo-EM structure of human glucose transporter GLUT4 bound to cytochalasin B in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytochalasin B, Solute carrier family 2, ... | | Authors: | Yuan, Y, Kong, F, Xu, H, Zhu, A, Yan, N, Yan, C. | | Deposit date: | 2022-01-30 | | Release date: | 2022-05-18 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Cryo-EM structure of human glucose transporter GLUT4.
Nat Commun, 13, 2022
|
|
7WSN
 
 | | Cryo-EM structure of human glucose transporter GLUT4 bound to cytochalasin B in detergent micelles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytochalasin B, Solute carrier family 2, ... | | Authors: | Yuan, Y, Kong, F, Xu, H, Zhu, A, Yan, N, Yan, C. | | Deposit date: | 2022-01-30 | | Release date: | 2022-05-18 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM structure of human glucose transporter GLUT4.
Nat Commun, 13, 2022
|
|
4GBY
 
 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-xylose | | Descriptor: | D-xylose-proton symporter, beta-D-xylopyranose, nonyl beta-D-glucopyranoside | | Authors: | Sun, L.F, Zeng, X, Yan, C.Y, Yan, N. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
4JA3
 
 | | Partially occluded inward open conformation of the xylose transporter XylE from E. coli | | Descriptor: | CADMIUM ION, D-xylose-proton symporter, LUTETIUM (III) ION | | Authors: | Quistgaard, E.M, Low, C, Moberg, P, Tresaugues, L, Nordlund, P. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for substrate transport in the GLUT-homology family of monosaccharide transporters.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JA4
 
 | | Inward open conformation of the xylose transporter XylE from E. coli | | Descriptor: | CADMIUM ION, D-xylose-proton symporter | | Authors: | Quistgaard, E.M, Low, C, Moberg, P, Tresaugues, L, Nordlund, P. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural basis for substrate transport in the GLUT-homology family of monosaccharide transporters.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4GC0
 
 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to 6-bromo-6-deoxy-D-glucose | | Descriptor: | 6-bromo-6-deoxy-beta-D-glucopyranose, D-xylose-proton symporter, nonyl beta-D-glucopyranoside | | Authors: | Yan, N, Sun, L.F, Zeng, X, Yan, C.Y. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
7D5Q
 
 | | Structure of NorC transporter (K398A mutant) in an outward-open conformation in complex with a single-chain Indian camelid antibody | | Descriptor: | Drug transporter, putative, ICab, ... | | Authors: | Kumar, S, Athreya, A, Penmatsa, A. | | Deposit date: | 2020-09-27 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis of inhibition of a transporter from Staphylococcus aureus, NorC, through a single-domain camelid antibody.
Commun Biol, 4, 2021
|
|
7D5P
 
 | | Structure of NorC transporter in an outward-open conformation in complex with a single-chain Indian camelid antibody | | Descriptor: | Drug transporter, putative, ICab, ... | | Authors: | Kumar, S, Athreya, A, Penmatsa, A. | | Deposit date: | 2020-09-27 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis of inhibition of a transporter from Staphylococcus aureus, NorC, through a single-domain camelid antibody.
Commun Biol, 4, 2021
|
|
6E9N
 
 | |
5EQH
 
 | | Human GLUT1 in complex with inhibitor (2~{S})-3-(2-bromophenyl)-2-[2-(4-methoxyphenyl)ethanoylamino]-~{N}-[(1~{S})-1-phenylethyl]propanamide | | Descriptor: | (2~{S})-3-(2-bromophenyl)-2-[2-(4-methoxyphenyl)ethanoylamino]-~{N}-[(1~{S})-1-phenylethyl]propanamide, Solute carrier family 2, facilitated glucose transporter member 1 | | Authors: | Kapoor, K, Finer-Moore, J, Pedersen, B.P, Caboni, L, Waight, A.B, Hillig, R, Bringmann, P, Heisler, I, Muller, T, Siebeneicher, H, Stroud, R.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Mechanism of inhibition of human glucose transporter GLUT1 is conserved between cytochalasin B and phenylalanine amides.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7SP5
 
 | | Crystal Structure of a Eukaryotic Phosphate Transporter | | Descriptor: | PHOSPHATE ION, Phosphate transporter, nonyl beta-D-glucopyranoside | | Authors: | Stroud, R.M, Pedersen, B.P, Kumar, H, Waight, A.B, Risenmay, A.J, Roe-Zurz, Z, Chau, B.H, Schlessinger, A, Bonomi, M, Harries, W, Sali, A, Johri, A.K, Finer-Moore, J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a eukaryotic phosphate transporter.
Nature, 496, 2013
|
|
