7DBK
 
 | | Crystal structure of human LDHB in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, L-lactate dehydrogenase B chain | | Authors: | Sogabe, S, Miwa, M. | | Deposit date: | 2020-10-20 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Identification of the first highly selective inhibitor of human lactate dehydrogenase B.
Sci Rep, 11, 2021
|
|
2J5R
 
 | | 2.25 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui after second radiation burn (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-19 | | Release date: | 2006-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
2J5Q
 
 | | 2.15 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui after first radiation burn (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-19 | | Release date: | 2006-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
5W8K
 
 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 29 and NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{3-(3,4-difluorophenyl)-5-hydroxy-4-[(4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, GLYCEROL, ... | | Authors: | Lukacs, C.M, Dranow, D.M. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
6Q13
 
 | | CRYSTAL STRUCTURE OF LDHA IN COMPLEX WITH COMPOUND NCGC00420737-09 AT 2.00 A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-[5-(cyclopropylmethyl)-4-[(3-fluoro-4-sulfamoylphenyl)methyl]-3-{3-[(5-methylthiophen-2-yl)ethynyl]phenyl}-1H-pyrazol-1-yl]-1,3-thiazole-4-carboxylic acid, ... | | Authors: | Davies, D.R, Dranow, D.M. | | Deposit date: | 2019-08-02 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyrazole-Based Lactate Dehydrogenase Inhibitors with Optimized Cell Activity and Pharmacokinetic Properties.
J.Med.Chem., 63, 2020
|
|
2J5K
 
 | | 2.0 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-18 | | Release date: | 2006-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
5W8I
 
 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 23 and Zinc | | Descriptor: | 2-[3-(3,4-difluorophenyl)-5-hydroxy-1H-pyrazol-1-yl]-1,3-thiazole-4-carboxylic acid, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Lukacs, C.M, Abendroth, J. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
6SBU
 
 | | X-ray Structure of Human LDHA with an Allosteric Inhibitor (Compound 3) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-[[4-[(5-chloranylthiophen-2-yl)carbonylamino]-1,3-bis(oxidanylidene)isoindol-2-yl]methyl]benzoic acid, L-lactate dehydrogenase A chain | | Authors: | Friberg, A, Puetter, V, Nguyen, D, Rehwinkel, H. | | Deposit date: | 2019-07-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural Evidence for Isoform-Selective Allosteric Inhibition of Lactate Dehydrogenase A.
Acs Omega, 5, 2020
|
|
1MLD
 
 | | REFINED STRUCTURE OF MITOCHONDRIAL MALATE DEHYDROGENASE FROM PORCINE HEART AND THE CONSENSUS STRUCTURE FOR DICARBOXYLIC ACID OXIDOREDUCTASES | | Descriptor: | CITRIC ACID, MALATE DEHYDROGENASE | | Authors: | Gleason, W.B, Fu, Z, Birktoft, J.J, Banaszak, L.J. | | Deposit date: | 1994-01-24 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Refined crystal structure of mitochondrial malate dehydrogenase from porcine heart and the consensus structure for dicarboxylic acid oxidoreductases.
Biochemistry, 33, 1994
|
|
6SBV
 
 | | X-ray Structure of Human LDH-A with an Allosteric Inhibitor (Compound 7) | | Descriptor: | L-lactate dehydrogenase A chain, ~{N}-[3-[(7-nitrodibenzofuran-2-yl)sulfonylamino]phenyl]-1-oxidanyl-cyclopropane-1-carboxamide | | Authors: | Friberg, A, Puetter, V, Nguyen, D, Rehwinkel, H. | | Deposit date: | 2019-07-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Evidence for Isoform-Selective Allosteric Inhibition of Lactate Dehydrogenase A.
Acs Omega, 5, 2020
|
|
5W8L
 
 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 59 and NADH | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{3-([1,1'-biphenyl]-3-yl)-5-(cyclopropylmethyl)-4-[(4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, ... | | Authors: | Davies, D.R, Dranow, D.M. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
2FN7
 
 | |
5W8H
 
 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 11 | | Descriptor: | 2-[3-(4-fluorophenyl)-5-(trifluoromethyl)-1H-pyrazol-1-yl]-1,3-thiazole-4-carboxylic acid, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Lukacs, C.M, Dranow, D.M. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
1PZH
 
 | | T.gondii LDH1 ternary complex with NAD and oxalate | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXALATE ION, lactate dehydrogenase | | Authors: | Kavanagh, K.L, Elling, R.A, Wilson, D.K. | | Deposit date: | 2003-07-10 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Toxoplasma gondii LDH1: Active-Site Differences from Human Lactate Dehydrogenases and the Structural Basis for Efficient APAD+ Use.
Biochemistry, 43, 2004
|
|
1PZE
 
 | | T.gondii LDH1 apo form | | Descriptor: | lactate dehydrogenase | | Authors: | Kavanagh, K.L, Elling, R.A, Wilson, D.K. | | Deposit date: | 2003-07-10 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Toxoplasma gondii LDH1: Active-Site Differences from Human Lactate Dehydrogenases and the Structural Basis for Efficient APAD+ Use.
Biochemistry, 43, 2004
|
|
1PZG
 
 | |
1PZF
 
 | | T.gondii LDH1 ternary complex with APAD+ and oxalate | | Descriptor: | 3-ACETYLPYRIDINE ADENINE DINUCLEOTIDE, OXALATE ION, lactate dehydrogenase | | Authors: | Kavanagh, K.L, Elling, R.A, Wilson, D.K. | | Deposit date: | 2003-07-10 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Toxoplasma gondii LDH1: Active-Site Differences from Human Lactate Dehydrogenases and the Structural Basis for Efficient APAD+ Use.
Biochemistry, 43, 2004
|
|
1D3A
 
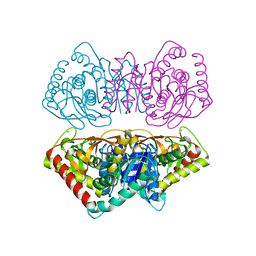 | | CRYSTAL STRUCTURE OF THE WILD TYPE HALOPHILIC MALATE DEHYDROGENASE IN THE APO FORM | | Descriptor: | CHLORIDE ION, HALOPHILIC MALATE DEHYDROGENASE, SODIUM ION | | Authors: | Richard, S.B, Madern, D, Garcin, E, Zaccai, G. | | Deposit date: | 1999-09-28 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Halophilic adaptation: novel solvent protein interactions observed in the 2.9 and 2.6 A resolution structures of the wild type and a mutant of malate dehydrogenase from Haloarcula marismortui.
Biochemistry, 39, 2000
|
|
1CIV
 
 | | CHLOROPLAST NADP-DEPENDENT MALATE DEHYDROGENASE FROM FLAVERIA BIDENTIS | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-MALATE DEHYDROGENASE | | Authors: | Carr, P.D, Ashton, A.R, Verger, D, Ollis, D.L. | | Deposit date: | 1999-04-01 | | Release date: | 2000-04-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chloroplast NADP-malate dehydrogenase: structural basis of light-dependent regulation of activity by thiol oxidation and reduction.
Structure Fold.Des., 7, 1999
|
|
1CEQ
 
 | | CHLOROQUINE BINDS IN THE COFACTOR BINDING SITE OF PLASMODIUM FALCIPARUM LACTATE DEHYDROGENASE. | | Descriptor: | PROTEIN (L-LACTATE DEHYDROGENASE) | | Authors: | Read, J.A, Wilkinson, K.W, Tranter, R, Sessions, R.B, Brady, R.L. | | Deposit date: | 1999-03-10 | | Release date: | 1999-03-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chloroquine binds in the cofactor binding site of Plasmodium falciparum lactate dehydrogenase.
J.Biol.Chem., 274, 1999
|
|
1CET
 
 | | CHLOROQUINE BINDS IN THE COFACTOR BINDING SITE OF PLASMODIUM FALCIPARUM LACTATE DEHYDROGENASE. | | Descriptor: | N4-(7-CHLORO-QUINOLIN-4-YL)-N1,N1-DIETHYL-PENTANE-1,4-DIAMINE, PROTEIN (L-LACTATE DEHYDROGENASE) | | Authors: | Read, J.A, Wilkinson, K.W, Tranter, R, Sessions, R.B, Brady, R.L. | | Deposit date: | 1999-03-10 | | Release date: | 1999-03-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Chloroquine binds in the cofactor binding site of Plasmodium falciparum lactate dehydrogenase.
J.Biol.Chem., 274, 1999
|
|
1EMD
 
 | |
1EZ4
 
 | | CRYSTAL STRUCTURE OF NON-ALLOSTERIC L-LACTATE DEHYDROGENASE FROM LACTOBACILLUS PENTOSUS AT 2.3 ANGSTROM RESOLUTION | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Uchikoba, H, Fushinobu, S, Wakagi, T, Konno, M, Taguchi, H, Matsuzawa, H. | | Deposit date: | 2000-05-10 | | Release date: | 2001-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of non-allosteric L-lactate dehydrogenase from Lactobacillus pentosus at 2.3 A resolution: specific interactions at subunit interfaces.
Proteins, 46, 2002
|
|
1GV1
 
 | | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases | | Descriptor: | MALATE DEHYDROGENASE | | Authors: | Dalhus, B, Sarinen, M, Sauer, U.H, Eklund, P, Johansson, K, Karlsson, A, Ramaswamy, S, Bjork, A, Synstad, B, Naterstad, K, Sirevag, R, Eklund, H. | | Deposit date: | 2002-02-04 | | Release date: | 2002-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases
J.Mol.Biol., 318, 2002
|
|
1GUY
 
 | | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases | | Descriptor: | CADMIUM ION, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dalhus, B, Sarinen, M, Sauer, U.H, Eklund, P, Johansson, K, Karlsson, A, Ramaswamy, S, Bjork, A, Synstad, B, Naterstad, K, Sirevag, R, Eklund, H. | | Deposit date: | 2002-02-04 | | Release date: | 2002-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases
J.Mol.Biol., 318, 2002
|
|
