3AVW
 
 | | Structure of viral RNA polymerase complex 4 | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVX
 
 | | Structure of viral RNA polymerase complex 5 | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVY
 
 | | Structure of viral RNA polymerase complex 6 | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3B78
 
 | | Structure of the eEF2-ExoA(R551H)-NAD+ complex | | Descriptor: | Elongation factor 2, Exotoxin A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jorgensen, R, Merrill, A.R. | | Deposit date: | 2007-10-30 | | Release date: | 2008-06-17 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The nature and character of the transition state for the ADP-ribosyltransferase reaction.
Embo Rep., 9, 2008
|
|
3B82
 
 | | Structure of the eEF2-ExoA(E546H)-NAD+ complex | | Descriptor: | Elongation factor 2, Exotoxin A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jorgensen, R, Merrill, A.R. | | Deposit date: | 2007-10-31 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The nature and character of the transition state for the ADP-ribosyltransferase reaction.
Embo Rep., 9, 2008
|
|
3B8H
 
 | | Structure of the eEF2-ExoA(E546A)-NAD+ complex | | Descriptor: | Elongation factor 2, Exotoxin A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jorgensen, R, Merrill, A.R. | | Deposit date: | 2007-11-01 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The nature and character of the transition state for the ADP-ribosyltransferase reaction.
Embo Rep., 9, 2008
|
|
3CB4
 
 | | The Crystal Structure of LepA | | Descriptor: | GTP-binding protein lepA | | Authors: | Evans, R.N, Blaha, G, Bailey, S, Steitz, T.A. | | Deposit date: | 2008-02-21 | | Release date: | 2008-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of LepA, the ribosomal back translocase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CW2
 
 | | Crystal structure of the intact archaeal translation initiation factor 2 from Sulfolobus solfataricus . | | Descriptor: | Translation initiation factor 2 subunit alpha, Translation initiation factor 2 subunit beta, Translation initiation factor 2 subunit gamma | | Authors: | Stolboushkina, E.A, Nikonov, S.V, Nikulin, A.D, Blaesi, U, Manstein, D.J, Fedorov, R.V, Garber, M.B, Nikonov, O.S. | | Deposit date: | 2008-04-21 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the intact archaeal translation initiation factor 2 demonstrates very high conformational flexibility in the alpha- and beta-subunits.
J.Mol.Biol., 382, 2008
|
|
3DEG
 
 | | Complex of elongating Escherichia coli 70S ribosome and EF4(LepA)-GMPPNP | | Descriptor: | 30S RNA helix 14, 30S RNA helix 8, 30S ribosomal protein S12, ... | | Authors: | Connell, S.R, Topf, M, Qin, Y, Wilson, D.N, Mielke, T, Fucini, P, Nierhaus, K.H, Spahn, C.M.T. | | Deposit date: | 2008-06-10 | | Release date: | 2008-08-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (10.9 Å) | | Cite: | A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation
Nat.Struct.Mol.Biol., 15, 2008
|
|
3DNY
 
 | |
3I1F
 
 | | Gamma-subunit of the translation initiation factor 2 from S. solfataricus in complex with Gpp(CH2)p | | Descriptor: | PHOSPHATE ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Translation initiation factor 2 subunit gamma | | Authors: | Stolboushkina, E.A, Nikonov, S.V, Nikulin, A.D, Blaesi, U, Garber, M.B, Nikonov, O.S. | | Deposit date: | 2009-06-26 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 5-prime-triphosphate end of mRNA recognizes the second GTP-binding site on the gamma-subunit of the archaeal translation initiation factor 2
To be Published
|
|
3IZP
 
 | |
3IZQ
 
 | | Structure of the Dom34-Hbs1-GDPNP complex bound to a translating ribosome | | Descriptor: | Elongation factor 1 alpha-like protein, Protein DOM34 | | Authors: | Becker, T, Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Sieber, H, Abdel Motaal, B, Mielke, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2010-11-30 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3IZY
 
 | | Mammalian mitochondrial translation initiation factor 2 | | Descriptor: | Translation initiation factor IF-2, mitochondrial, tRNA-Phe | | Authors: | Yassin, A.S, Haque, E, Datta, P.P, Elmore, K, Banavali, N.K, Spremulli, L.L, Agrawal, R.K. | | Deposit date: | 2011-01-19 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Insertion domain within mammalian mitochondrial translation initiation factor 2 serves the role of eubacterial initiation factor 1
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3J0E
 
 | | Models for the T. thermophilus ribosome recycling factor and the E. coli elongation factor G bound to the E. coli post-termination complex | | Descriptor: | 30S ribosomal protein S12, Elongation factor G, Ribosome-recycling factor, ... | | Authors: | Yokoyama, T, Shaikh, T.R, Iwakura, N, Kaji, H, Kaji, A, Agrawal, R.K. | | Deposit date: | 2011-06-29 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | Structural insights into initial and intermediate steps of the ribosome-recycling process.
Embo J., 31, 2012
|
|
3J25
 
 | | Structural basis for TetM-mediated tetracycline resistance | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Tetracycline resistance protein tetM | | Authors: | Doenhoefer, A, Franckenberg, S, Wickles, S, Berninghausen, O, Beckmann, R, Wilson, D.N. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structural basis for TetM-mediated tetracycline resistance.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3J4J
 
 | | Model of full-length T. thermophilus Translation Initiation Factor 2 refined against its cryo-EM density from a 30S Initiation Complex map | | Descriptor: | Translation initiation factor IF-2 | | Authors: | Simonetti, A, Marzi, S, Billas, I.M.L, Tsai, A, Fabbretti, A, Myasnikov, A, Roblin, P, Vaiana, A.C, Hazemann, I, Eiler, D, Steitz, T.A, Puglisi, J.D, Gualerzi, C.O, Klaholz, B.P. | | Deposit date: | 2013-08-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Involvement of protein IF2 N domain in ribosomal subunit joining revealed from architecture and function of the full-length initiation factor.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3J5Y
 
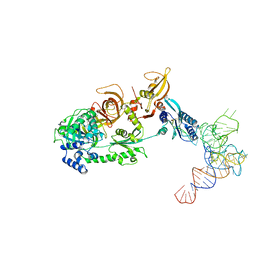 | | Structure of the mammalian ribosomal pre-termination complex associated with eRF1-eRF3-GDPNP | | Descriptor: | 5'-R(*AP*UP*UP*GP*UP*AP*AP*AP*AP*A)-3', Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, Eukaryotic peptide chain release factor subunit 1, ... | | Authors: | des Georges, A, Hashem, Y, Unbehaun, A, Grassucci, R.A, Taylor, D, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-25 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Structure of the mammalian ribosomal pre-termination complex associated with eRF1*eRF3*GDPNP.
Nucleic Acids Res., 42, 2014
|
|
3J81
 
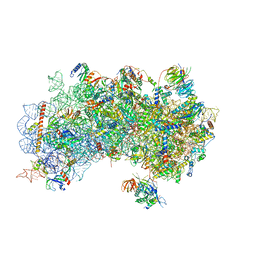 | | CryoEM structure of a partial yeast 48S preinitiation complex | | Descriptor: | 18S rRNA, MAGNESIUM ION, METHIONINE, ... | | Authors: | Hussain, T, Llacer, J.L, Fernandez, I.S, Savva, C.G, Ramakrishnan, V. | | Deposit date: | 2014-08-29 | | Release date: | 2014-11-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural changes enable start codon recognition by the eukaryotic translation initiation complex.
Cell(Cambridge,Mass.), 159, 2014
|
|
3J9Y
 
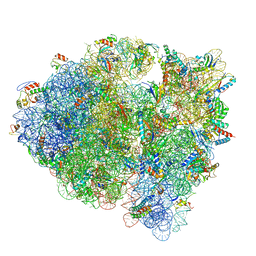 | | Cryo-EM structure of tetracycline resistance protein TetM bound to a translating E.coli ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Arenz, S, Nguyen, F, Beckmann, R, Wilson, D.N. | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-15 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the tetracycline resistance protein TetM in complex with a translating ribosome at 3.9- angstrom resolution.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3J9Z
 
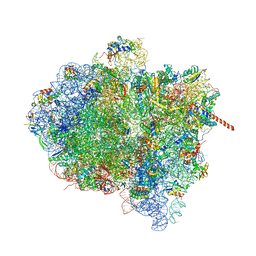 | | Activation of GTP Hydrolysis in mRNA-tRNA Translocation by Elongation Factor G | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Li, W, Liu, Z, Koripella, R.K, Langlois, R, Sanyal, S, Frank, J. | | Deposit date: | 2015-03-27 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G.
Sci Adv, 1, 2015
|
|
3JA1
 
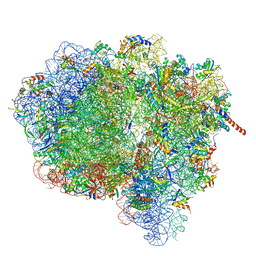 | | Activation of GTP Hydrolysis in mRNA-tRNA Translocation by Elongation Factor G | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Li, W, Liu, Z, Koripella, R.K, Langlois, R, Sanyal, S, Frank, J. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G.
Sci Adv, 1, 2015
|
|
3JAP
 
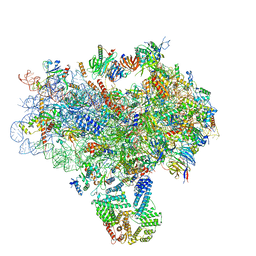 | | Structure of a partial yeast 48S preinitiation complex in closed conformation | | Descriptor: | 18S rRNA, MAGNESIUM ION, METHIONINE, ... | | Authors: | Llacer, J.L, Hussain, T, Ramakrishnan, V. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Conformational Differences between Open and Closed States of the Eukaryotic Translation Initiation Complex.
Mol.Cell, 59, 2015
|
|
3JB9
 
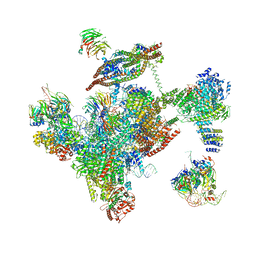 | | Cryo-EM structure of the yeast spliceosome at 3.6 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yan, C, Hang, J, Wan, R, Huang, M, Wong, C, Shi, Y. | | Deposit date: | 2015-08-09 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a yeast spliceosome at 3.6-angstrom resolution
Science, 349, 2015
|
|
3JCD
 
 | | Structure of Escherichia coli EF4 in posttranslocational ribosomes (Post EF4) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
