6KNS
 
 | | Crystal structure of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis (space group I4122) | | 分子名称: | CALCIUM ION, Putative metal-dependent hydrolase, ZINC ION | | 著者 | Na, H.W, Namgung, B, Song, W.S, Yoon, S.I. | | 登録日 | 2019-08-07 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural and biochemical analyses of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
6KNT
 
 | | Crystal structure of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis (space group P4332) | | 分子名称: | Putative metal-dependent hydrolase, ZINC ION | | 著者 | Na, H.W, Namgung, B, Song, W.S, Yoon, S.I. | | 登録日 | 2019-08-07 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and biochemical analyses of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
5WU7
 
 | |
5JOC
 
 | | Crystal structure of the S61A mutant of AmpC BER | | 分子名称: | Beta-lactamase, CITRIC ACID | | 著者 | Na, J.H, An, Y.J, Cha, S.S. | | 登録日 | 2016-05-02 | | 公開日 | 2017-05-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis for the extended substrate spectrum of AmpC BER and structure-guided discovery of the inhibition activity of citrate against the class C beta-lactamases AmpC BER and CMY-10.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5JXE
 
 | |
1SG8
 
 | | Crystal structure of the procoagulant fast form of thrombin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, thrombin | | 著者 | Pineda, A.O, Carrell, C.J, Bush, L.A, Prasad, S, Caccia, S, Chen, Z.W, Mathews, F.S, Di Cera, E. | | 登録日 | 2004-02-23 | | 公開日 | 2004-06-08 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular dissection of na+ binding to thrombin.
J.Biol.Chem., 279, 2004
|
|
2E30
 
 | |
8V9H
 
 | | GES-5-NA-1-157 complex | | 分子名称: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | 著者 | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | 登録日 | 2023-12-08 | | 公開日 | 2024-04-03 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Restricted Rotational Flexibility of the C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Leads to Potent Inhibition of the GES-5 Carbapenemase.
Acs Infect Dis., 10, 2024
|
|
4UET
 
 | | Diversity in the structures and ligand binding sites among the fatty acid and retinol binding proteins of nematodes revealed by Na-FAR-1 from Necator americanus | | 分子名称: | NEMATODE FATTY ACID RETINOID BINDING PROTEIN | | 著者 | Rey-Burusco, M.F, Ibanez Shimabukuro, M, Griffiths, K, Cooper, A, Kennedy, M.W, Corsico, B, Smith, B.O, Griffiths, K. | | 登録日 | 2014-12-18 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Diversity in the Structures and Ligand Binding Sites of Nematode Fatty Acid and Retinol Binding Proteins Revealed by Na-Far-1 from Necator Americanus.
Biochem.J., 471, 2015
|
|
4U9S
 
 | |
4U9O
 
 | | Crystal structure of NqrA from Vibrio cholerae | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Na(+)-translocating NADH-quinone reductase subunit A | | 著者 | Fritz, G. | | 登録日 | 2014-08-06 | | 公開日 | 2014-12-03 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of the V. cholerae Na+-pumping NADH:quinone oxidoreductase.
Nature, 516, 2014
|
|
4UAJ
 
 | | Crystal structure of NqrF in hexagonal space group | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit F, SULFATE ION | | 著者 | Fritz, G. | | 登録日 | 2014-08-10 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.7019 Å) | | 主引用文献 | Structure of the V. cholerae Na+-pumping NADH:quinone oxidoreductase.
Nature, 516, 2014
|
|
4US4
 
 | | Crystal Structure of the Bacterial NSS Member MhsT in an Occluded Inward-Facing State (lipidic cubic phase form) | | 分子名称: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, SODIUM ION, ... | | 著者 | Malinauskaite, L, Quick, M, Reinhard, L, Lyons, J.A, Yano, H, Javitch, J.A, Nissen, P. | | 登録日 | 2014-07-02 | | 公開日 | 2014-09-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A Mechanism for Intracellular Release of Na+ by Neurotransmitter/Sodium Symporters
Nat.Struct.Mol.Biol., 21, 2014
|
|
4US3
 
 | | Crystal Structure of the bacterial NSS member MhsT in an Occluded Inward-Facing State | | 分子名称: | DODECYL-ALPHA-D-MALTOSIDE, SODIUM ION, TRANSPORTER, ... | | 著者 | Malinauskaite, L, Quick, M, Reinhard, L, Lyons, J.A, Yano, H, Javitch, J.A, Nissen, P. | | 登録日 | 2014-07-02 | | 公開日 | 2014-09-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.098 Å) | | 主引用文献 | A Mechanism for Intracellular Release of Na+ by Neurotransmitter/Sodium Symporters
Nat.Struct.Mol.Biol., 21, 2014
|
|
8A1X
 
 | | Sodium pumping NADH-quinone oxidoreductase with inhibitor DQA | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | 登録日 | 2022-06-02 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1Y
 
 | | Sodium pumping NADH-quinone oxidoreductase with inhibitor HQNO | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, DODECYL-BETA-D-MALTOSIDE, ... | | 著者 | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | 登録日 | 2022-06-02 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4XCP
 
 | | Fatty Acid and Retinol binding protein Na-FAR-1 from Necator americanus | | 分子名称: | Nematode fatty acid retinoid binding protein, PALMITIC ACID | | 著者 | Gabrielsen, M, Rey-Burusco, M.F, Ibanez-Shimabukuro, M, Griffiths, K, Kennedy, M.W, Corsico, B, Smith, B.O. | | 登録日 | 2014-12-18 | | 公開日 | 2015-09-16 | | 最終更新日 | 2017-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Diversity in the structures and ligand-binding sites of nematode fatty acid and retinol-binding proteins revealed by Na-FAR-1 from Necator americanus.
Biochem.J., 471, 2015
|
|
7QTV
 
 | | Beryllium fluoride form of the Na+,K+-ATPase (E2-BeFx) | | 分子名称: | 1-O-decanoyl-beta-D-tagatofuranosyl beta-D-allopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Fruergaard, M.U, Dach, I, Andersen, J.L, Ozol, M, Shahsavar, A, Quistgaard, E.M, Poulsen, H, Fedosova, N.U, Nissen, P. | | 登録日 | 2022-01-16 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (4.05 Å) | | 主引用文献 | The Na + ,K + -ATPase in complex with beryllium fluoride mimics an ATPase phosphorylated state.
J.Biol.Chem., 298, 2022
|
|
7RPK
 
 | | Cryo-EM structure of murine Dispatched in complex with Sonic hedgehog | | 分子名称: | (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | 登録日 | 2021-08-03 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7R63
 
 | |
7D91
 
 | | Crystal Structure of the Na+,K+-ATPase in the E2P state with bound Mg2+ (P4(3)2(1)2 symmetry) | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kanai, R, Cornelius, F, Ogawa, H, Motoyama, K, Vilsen, B, Toyoshima, C. | | 登録日 | 2020-10-12 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7D92
 
 | | Crystal Structure of the Na+,K+-ATPase in the E2P state with bound Mg2+ and anthroylouabain (P4(3)2(1)2 symmetry) | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kanai, R, Cornelius, F, Ogawa, H, Motoyama, K, Vilsen, B, Toyoshima, C. | | 登録日 | 2020-10-12 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RPI
 
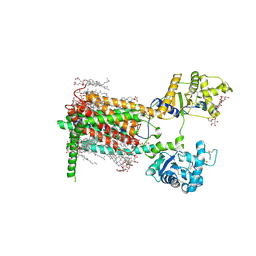 | | Cryo-EM structure of murine Dispatched 'T' conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | 著者 | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | 登録日 | 2021-08-03 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7RPH
 
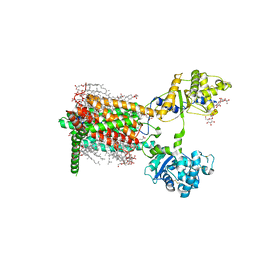 | | Cryo-EM structure of murine Dispatched 'R' conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | 著者 | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | 登録日 | 2021-08-03 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
8I5Y
 
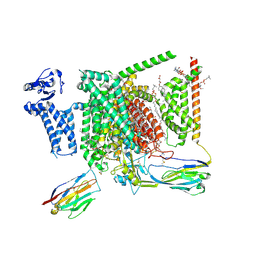 | | Structure of human Nav1.7 in complex with vixotrigine | | 分子名称: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | 著者 | Wu, Q.R, Yan, N. | | 登録日 | 2023-01-26 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
