8JT3
 
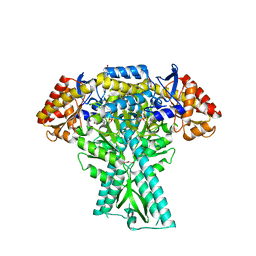 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with amino donor L-Arg | | Descriptor: | (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, ACETATE ION, CrmG, ... | | Authors: | Su, K, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2023-06-21 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Co-crystal structure provides insights on transaminase CrmG recognition amino donor L-Arg
Biochem.Biophys.Res.Commun., 675, 2023
|
|
8JNF
 
 | | The cryo-EM structure of the RAD51 filament bound to the nucleosome | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.91 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site
Nature, 628, 2024
|
|
8JNE
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome without the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site
Nature, 628, 2024
|
|
8JND
 
 | | The cryo-EM structure of the nonameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site
Nature, 628, 2024
|
|
8JLD
 
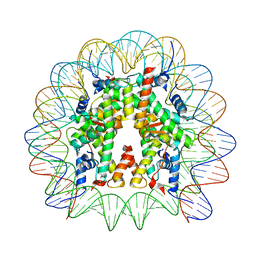 | | Cryo-EM structure of the 145 bp human nucleosome containing acetylated H3 tail | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
8JLB
 
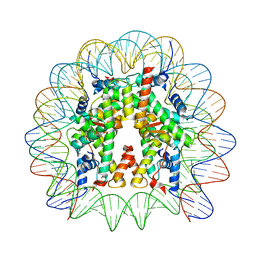 | | Cryo-EM structure of the 145 bp human nucleosome containing H3.2 C110A mutant | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
8JLA
 
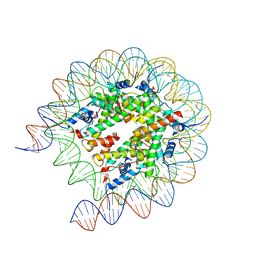 | | Cryo-EM structure of the human nucleosome lacking N-terminal region of H2A, H2B, H3, and H4 | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
8JL9
 
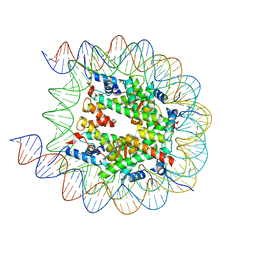 | | Cryo-EM structure of the human nucleosome with scFv | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
8JHO
 
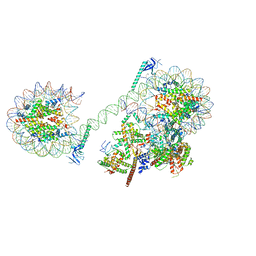 | |
8JHG
 
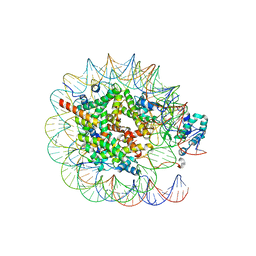 | | Native SUV420H1 bound to 167-bp nucleosome | | Descriptor: | DNA (160-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Lin, F, Li, W. | | Deposit date: | 2023-05-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis of nucleosomal H4K20 recognition and methylation by SUV420H1 methyltransferase.
Cell Discov, 9, 2023
|
|
8JHF
 
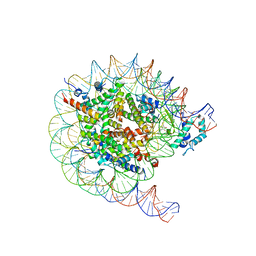 | | Native SUV420H1 bound to 167-bp nucleosome | | Descriptor: | DNA (160-MER), Histone H2A.Z, Histone H2B type 1-K, ... | | Authors: | Lin, F, Li, W. | | Deposit date: | 2023-05-23 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis of nucleosomal H4K20 recognition and methylation by SUV420H1 methyltransferase.
Cell Discov, 9, 2023
|
|
8JH4
 
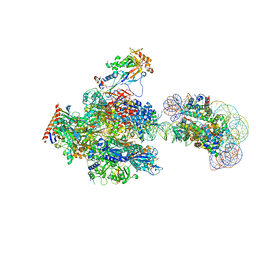 | | RNA polymerase II elongation complex containing 60 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
8JH3
 
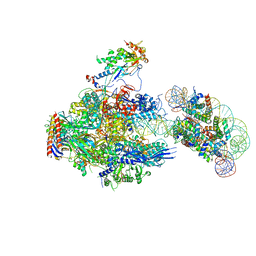 | | RNA polymerase II elongation complex containing 40 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
8JH2
 
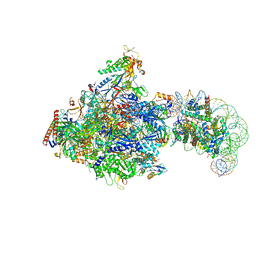 | | RNA polymerase II elongation complex bound with Elf1, Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (218-MER), DNA (40-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
8JE4
 
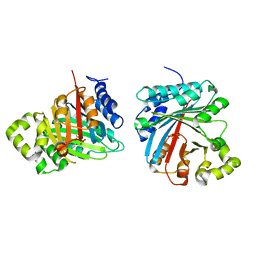 | | Crystal structure of LimF prenyltransferase (H239G/W273T mutant) bound with the thiodiphosphate moiety of farnesyl S-thiolodiphosphate (FSPP) | | Descriptor: | MAGNESIUM ION, TRIHYDROGEN THIODIPHOSPHATE, prenyltransferase, ... | | Authors: | Hamada, K, Oguni, A, Zhang, Y, Satake, M, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Switching Prenyl Donor Specificities of Cyanobactin Prenyltransferases.
J.Am.Chem.Soc., 145, 2023
|
|
8JC4
 
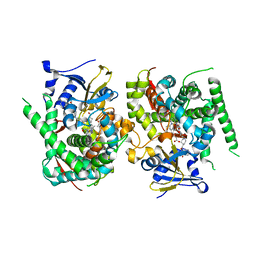 | | Crystal structure of the P450 BM3 heme domain mutant F87A-T268V in complex with Pyd-Pid-Phe and hydroxylamine | | Descriptor: | 1-pyridin-4-ylpiperidine-4-carboxylic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2023-05-10 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | C(sp3)-H hydroxylation of Broad-Spectrum Alkanes Catalyzed by an Artificial P450 Peroxygenase Driven by omega-Pyridyl Fatty Acyl Amino Acids.
Mol Catal, 550, 2023
|
|
8JC3
 
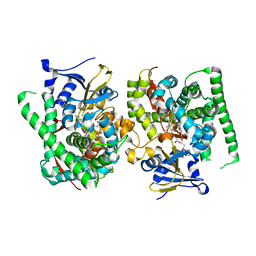 | | Crystal structure of the P450 BM3 heme domain mutant F87A-T268V in complex with Pyd-N-C4-Phe and hydroxylamine | | Descriptor: | 4-(pyridin-4-ylamino)butanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2023-05-10 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | C(sp3)-H hydroxylation of Broad-Spectrum Alkanes Catalyzed by an Artificial P450 Peroxygenase Driven by omega-Pyridyl Fatty Acyl Amino Acids.
Mol Catal, 550, 2023
|
|
8J6T
 
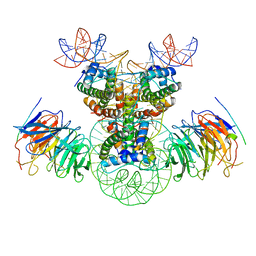 | | Cryo-EM structure of the double CAF-1 bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8J6S
 
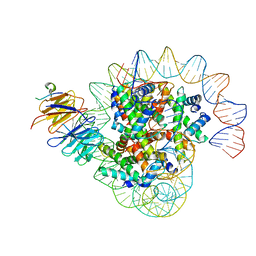 | | Cryo-EM structure of the single CAF-1 bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8J5M
 
 | |
8J24
 
 | | Cryo-EM structure of FFAR2 complex bound with acetic acid | | Descriptor: | ACETATE ION, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Tang, W, Sun, X, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J07
 
 | |
8IYJ
 
 | | Cryo-EM structure of the 48-nm repeat doublet microtubule from mouse sperm | | Descriptor: | Cilia and flagella-associated protein 77, Cilia- and flagella-associated protein 107, Cilia- and flagella-associated protein 141, ... | | Authors: | Zhou, L.N, Gui, M, Wu, J.P. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of sperm flagellar doublet microtubules expand the genetic spectrum of male infertility.
Cell, 186, 2023
|
|
8IYC
 
 | | Structure insight into substrate recognition and catalysis by feruloyl esterase from Aspergillus sydowii | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the molecular mechanisms of substrate recognition and hydrolysis by feruloyl esterase from Aspergillus sydowii.
Int.J.Biol.Macromol., 253, 2023
|
|
8IYB
 
 | | Structure insight into substrate recognition and catalysis by feruloyl esterase from Aspergillus sydowii | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ... | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the molecular mechanisms of substrate recognition and hydrolysis by feruloyl esterase from Aspergillus sydowii.
Int.J.Biol.Macromol., 253, 2023
|
|
