7K2R
 
 | | Kelch domain of human KEAP1 bound to Nrf2-based cyclic peptide, c[LhA-DEETGE] | | Descriptor: | B3A-ASP-PRO-GLU-THR-GLY-GLU, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K2Q
 
 | | Kelch domain of human KEAP1 bound to Nrf2 cyclic peptide, c[Ahx-DPETGE] | | Descriptor: | ACA-ASP-PRO-GLU-THR-GLY-GLU, Kelch-like ECH-associated protein 1 | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K29
 
 | | Kelch domain of human KEAP1 bound to Nrf2 peptide, LDEETGEAL | | Descriptor: | ACE-LEU-ASP-GLU-GLU-THR-GLY-GLU-ALA-LEU-NH2, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
6ZNC
 
 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human wild-type p53DBD bound to DNA and MQ: wt-DNA-MQ (I) | | Descriptor: | (2~{R})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, (2~{S})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, Cellular tumor antigen p53, ... | | Authors: | Rozenberg, H, Degtjarik, O, Diskin-Posner, Y, Shakked, Z. | | Deposit date: | 2020-07-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
5AFM
 
 | | alpha7-AChBP in complex with lobeline and fragment 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,5-dibromo-N-(3-hydroxypropyl)-1H-pyrrole-2-carboxamide, ACETYLCHOLINE-BINDING PROTEIN, ... | | Authors: | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7K98
 
 | | Preaminoacylation complex of M. tuberculosis PheRS with cognate precursor tRNA and 5'-O-(N-phenylalanyl)sulfamoyl-adenosine (F-AMS) | | Descriptor: | 5'-O-(L-phenylalanylsulfamoyl)adenosine, CHLORIDE ION, GLYCEROL, ... | | Authors: | Michalska, K, Chang, C, Jedrzejczak, R, Wower, J, Baragana, B, Forte, B, Gilbert, I.H, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-28 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation.
Nucleic Acids Res., 49, 2021
|
|
7K9M
 
 | | Crystal structure of the complex of M. tuberculosis PheRS with cognate precursor tRNA and 5'-O-(N-phenylalanyl)sulfamoyl-adenosine | | Descriptor: | 5'-O-(L-phenylalanylsulfamoyl)adenosine, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Michalska, K, Chang, C, Jedrzejczak, R, Wower, J, Baragana, B, Forte, B, Gilbert, I.H, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation.
Nucleic Acids Res., 49, 2021
|
|
5AFJ
 
 | | alpha7-AChBP in complex with lobeline and fragment 1 | | Descriptor: | (3S)-6-(4-bromophenyl)-3-hydroxy-1,3-dimethyl-2,3-dihydropyridin-4(1H)-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN, ... | | Authors: | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4DK8
 
 | | Crystal structure of LXR ligand binding domain in complex with partial agonist 5 | | Descriptor: | ACETATE ION, CALCIUM ION, N-methyl-N-(4-{(1S)-2,2,2-trifluoro-1-hydroxy-1-[1-(2-methoxyethyl)-1H-pyrrol-2-yl]ethyl}phenyl)benzenesulfonamide, ... | | Authors: | Piper, D.E, Kopecky, D.J, Xu, H. | | Deposit date: | 2012-02-03 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of a new binding mode for a series of liver X receptor agonists.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3WN4
 
 | | Crystal structure of human TLR8 in complex with DS-877 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-butylfuro[2,3-c]quinolin-4-amine, Toll-like receptor 8, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-based design of novel human Toll-like receptor 8 agonists.
Chemmedchem, 9, 2014
|
|
1V6X
 
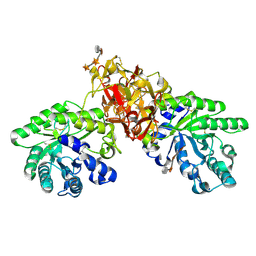 | | Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 3(3)-4-O-methyl-alpha-D-glucuronosyl-xylotriose | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ENDO-1,4-BETA-D-XYLANASE, beta-D-xylopyranose, ... | | Authors: | Fujimoto, Z, Kaneko, S, Kuno, A, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2003-12-04 | | Release date: | 2004-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86
J.Biol.Chem., 279, 2004
|
|
5J5I
 
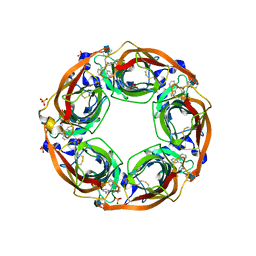 | | X-Ray Crystal Structure of Acetylcholine Binding Protein (AChBP) in Complex with 4-(2-amino-6-{bis[(pyridin-2-yl)methyl]amino}pyrimidin-4-yl)phenol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-amino-6-{bis[(pyridin-2-yl)methyl]amino}pyrimidin-4-yl)phenol, Acetylcholine-binding protein, ... | | Authors: | Kaczanowska, K, Harel, M, Camacho Hernandez, G.A, Taylor, P. | | Deposit date: | 2016-04-02 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.326 Å) | | Cite: | Substituted 2-Aminopyrimidines Selective for alpha 7-Nicotinic Acetylcholine Receptor Activation and Association with Acetylcholine Binding Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
5J5H
 
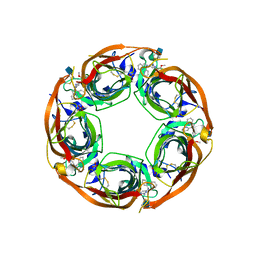 | | X-RAY STRUCTURE OF ACETYLCHOLINE BINDING PROTEIN (ACHBP) IN COMPLEX WITH 6-(2-methoxyphenyl)-N4,N4-bis[(pyridin-2-yl)methyl]pyrimidine-2,4-diamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(2-methoxyphenyl)-N~4~,N~4~-bis[(pyridin-2-yl)methyl]pyrimidine-2,4-diamine, Acetylcholine-binding protein, ... | | Authors: | Kaczanowska, K, Harel, M, Camacho Hernandez, A.G, Taylor, P. | | Deposit date: | 2016-04-02 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substituted 2-Aminopyrimidines Selective for alpha 7-Nicotinic Acetylcholine Receptor Activation and Association with Acetylcholine Binding Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
8TTQ
 
 | |
6S98
 
 | |
5J5G
 
 | | X-Ray Crystal Structure of Acetylcholine Binding Protein (AChBP) in Complex with 6-(4-methoxyphenyl)-N4,N4-bis[(pyridin-2-yl)methyl]pyrimidine-2,4-diamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(4-methoxyphenyl)-N~4~,N~4~-bis[(pyridin-2-yl)methyl]pyrimidine-2,4-diamine, Acetylcholine-binding protein, ... | | Authors: | Kaczanowska, K, Harel, M, Camacho Hernandez, A.G, Taylor, P. | | Deposit date: | 2016-04-02 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.036 Å) | | Cite: | Substituted 2-Aminopyrimidines Selective for alpha 7-Nicotinic Acetylcholine Receptor Activation and Association with Acetylcholine Binding Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
6S96
 
 | |
2ECU
 
 | | Crystal structure of flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, DODECAETHYLENE GLYCOL, flavin reductase (HpaC) of 4-hydroxyphenylacetate 3-monooxygnease | | Authors: | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-02-14 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
1G0F
 
 | |
2Z3G
 
 | | Crystal structure of blasticidin S deaminase (BSD) | | Descriptor: | Blasticidin-S deaminase, ZINC ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Kumasaka, T, Yamamoto, M, Furuichi, M, Nakasako, M, Kimura, M, Yamaguchi, I, Ueki, T. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc
J.Biol.Chem., 282, 2007
|
|
5AFH
 
 | | alpha7-AChBP in complex with lobeline | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN, NEURONAL ACETYLCHOLINE RECEPTOR SUBUNIT ALPHA-7, ... | | Authors: | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1YJK
 
 | | Reduced Peptidylglycine Alpha-Hydroxylating Monooxygenase (PHM) in a New Crystal Form | | Descriptor: | COPPER (II) ION, GLYCEROL, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Siebert, X, Eipper, B.A, Mains, R.E, Prigge, S.T, Blackburn, N.J, Amzel, L.M. | | Deposit date: | 2005-01-14 | | Release date: | 2005-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Catalytic Copper of Peptidylglycine alpha-Hydroxylating Monooxygenase also Plays a Critical Structural Role.
Biophys.J., 89, 2005
|
|
1YJL
 
 | | Reduced Peptidylglycine alpha-Hydroxylating Monooxygenase in a new crystal form | | Descriptor: | Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Siebert, X, Eipper, B.A, Mains, R.E, Prigge, S.T, Blackburn, N.J, Amzel, L.M. | | Deposit date: | 2005-01-14 | | Release date: | 2005-11-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The catalytic copper of Peptidylglycine alpha-Hydroxylating Monooxygenase also plays a critical structural role.
Biophys.J., 89, 2005
|
|
5J5F
 
 | | X-Ray Crystal Structure of Acetylcholine Binding Protein (AChBP) in Complex with N4,N4-bis[(pyridin-2-yl)methyl]-6-(thiophen-3-yl)pyrimidine-2,4-diamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Kaczanowska, K, Camacho Hernandez, G.A, Harel, M, Taylor, P. | | Deposit date: | 2016-04-02 | | Release date: | 2017-03-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Substituted 2-Aminopyrimidines Selective for alpha 7-Nicotinic Acetylcholine Receptor Activation and Association with Acetylcholine Binding Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
3SOX
 
 | | Structure of UHRF1 PHD finger in the free form | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Rajakumara, E, Patel, D.J. | | Deposit date: | 2011-06-30 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6501 Å) | | Cite: | PHD Finger Recognition of Unmodified Histone H3R2 Links UHRF1 to Regulation of Euchromatic Gene Expression.
Mol.Cell, 43, 2011
|
|
