7C5C
 
 | |
3JWA
 
 | | Crystal structure of L-methionine gamma-lyase from Citrobacter freundii with methionine phosphinate | | Descriptor: | (1-AMINO-3-METHYLSULFANYL-PROPYL)-PHOSPHINIC ACID, DI(HYDROXYETHYL)ETHER, Methionine gamma-lyase | | Authors: | Revtovish, S.V, Nikulin, A.D, Morozova, E.A, Demidkina, T.V. | | Deposit date: | 2009-09-18 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Three-dimensional structures of noncovalent complexes of Citrobacter freundii methionine gamma-lyase with substrates.
Biochemistry Mosc., 76, 2011
|
|
6S7S
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(2-Methoxy-5-(N-phenylsulfamoyl)phenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, ~{N}-[2-methoxy-5-(phenylsulfamoyl)phenyl]-2-(4-oxidanylidene-3~{H}-phthalazin-1-yl)ethanamide | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-05 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
9FBY
 
 | |
9FBX
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 5-(1-benzyl-4-chloro-1H-imidazol-2-yl)-1,3-dimethylpyridin-2(1H)-one | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 5-(1-benzyl-4-chloro-1H-imidazol-2-yl)-1,3-dimethylpyridin-2(1H)-one, ... | | Authors: | Chung, C. | | Deposit date: | 2024-05-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Structure- and Property-Based Optimization of Efficient Pan-Bromodomain and Extra Terminal Inhibitors to Identify Oral and Intravenous Candidate I-BET787.
J.Med.Chem., 67, 2024
|
|
6RP3
 
 | | Truncated Norcoclaurine synthase with reaction intermediate mimic | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-[[(2~{R})-2-phenylpropyl]amino]ethyl]benzene-1,2-diol, S-norcoclaurine synthase | | Authors: | Keep, N.H, Roddan, R, Sula, A. | | Deposit date: | 2019-05-13 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Acceptance and Kinetic Resolution of alpha-Methyl-Substituted Aldehydes by Norcoclaurine Synthases
Acs Catalysis, 2019
|
|
5ECD
 
 | | Structure of the Shigella flexneri VapC mutant D98N crystal form 1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, tRNA(fMet)-specific endonuclease VapC | | Authors: | Xu, K. | | Deposit date: | 2015-10-20 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of the active site architecture of the VapC toxin from Shigella flexneri.
Proteins, 84, 2016
|
|
8RVQ
 
 | | 20S proteasome from pre1-1 | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Mark, E, Ramos, P.C, Kayser, F, Hoeckendorff, J, Dohmen, R.J, Wendler, P. | | Deposit date: | 2024-02-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.02 Å) | | Cite: | Structural roles of Ump1 and beta-subunit propeptides in proteasome biogenesis.
Life Sci Alliance, 7, 2024
|
|
6UWU
 
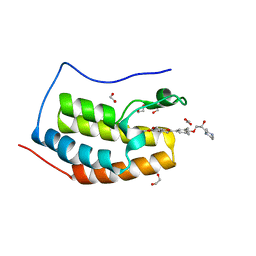 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor ZL0516 | | Descriptor: | 1,2-ETHANEDIOL, 2-{4-[(2R)-2-hydroxy-3-(4-methylpiperazin-1-yl)propoxy]-3,5-dimethylphenyl}-5,7-dimethoxy-4H-1-benzopyran-4-one, Bromodomain-containing protein 4 | | Authors: | Leonard, P.G, Joseph, S. | | Deposit date: | 2019-11-05 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Orally Bioavailable Chromone Derivatives as Potent and Selective BRD4 Inhibitors: Scaffold Hopping, Optimization, and Pharmacological Evaluation.
J.Med.Chem., 63, 2020
|
|
8EB3
 
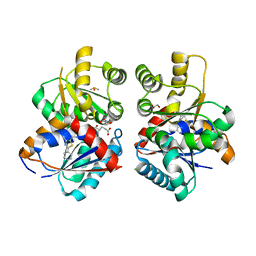 | |
8AY6
 
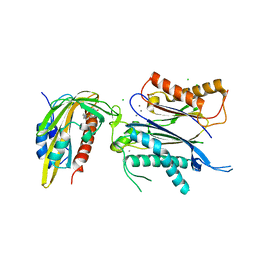 | | X-RAY CRYSTAL STRUCTURE OF THE CsPYL1(V112L, T135L,F137I, T153I, V168A)-SB-HAB1 TERNARY COMPLEX | | Descriptor: | 1,4-dimethyl-2-oxidanylidene-~{N}-(phenylmethyl)quinoline-6-sulfonamide, Abscisic acid receptor PYL1, CHLORIDE ION, ... | | Authors: | Infantes, L, Albert, A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-guided engineering of a receptor-agonist pair for inducible activation of the ABA adaptive response to drought.
Sci Adv, 9, 2023
|
|
8ABG
 
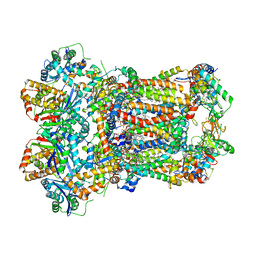 | | Complex III2 from Yarrowia lipolytica, oxidised with ferricyanide, c-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
5II2
 
 | | Crystal Structure of the fifth bromodomain of human polybromo (PB1) in complex with 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, CITRIC ACID, POTASSIUM ION, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S. | | Deposit date: | 2016-03-01 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Optimization of a Selective Ligand for the Switch/Sucrose Nonfermenting-Related Bromodomains of Polybromo Protein-1 by the Use of Virtual Screening and Hydration Analysis.
J.Med.Chem., 59, 2016
|
|
8WY1
 
 | | The structure of cyclization domain in cyclic beta-1,2-glucan synthase from Thermoanaerobacter italicus | | Descriptor: | Glycosyltransferase 36 | | Authors: | Tanaka, N, Saito, R, Kobayashi, K, Nakai, H, Kamo, S, Kuramochi, K, Taguchi, H, Nakajima, M, Masaike, T. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Functional and structural analysis of a cyclization domain in a cyclic beta-1,2-glucan synthase.
Appl.Microbiol.Biotechnol., 108, 2024
|
|
5L3O
 
 | | Crystal Structure of Human Carbonic Anhydrase II in Complex with a Quinoline Oligoamide Foldamer | | Descriptor: | Aromatic foldamer, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Jewginski, M, Langlois d'Estaintot, B, Granier, T, Huc, Y. | | Deposit date: | 2016-05-24 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Self-Assembled Protein-Aromatic Foldamer Complexes with 2:3 and 2:2:1 Stoichiometries.
J. Am. Chem. Soc., 139, 2017
|
|
5IKK
 
 | | Structure of the histone deacetylase Clr3 | | Descriptor: | 1,2-ETHANEDIOL, Histone deacetylase clr3, MAGNESIUM ION, ... | | Authors: | Brugger, C, Schalch, T. | | Deposit date: | 2016-03-03 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SHREC Silences Heterochromatin via Distinct Remodeling and Deacetylation Modules.
Mol.Cell, 62, 2016
|
|
9FDJ
 
 | | Crystal structure of the NuoEF variant R66G (NuoF) from Aquifex aeolicus bound to NADH under anoxic conditions (short soak) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FIF
 
 | | Crystal Structure of NuoEF variant P228R(NuoF) from Aquifex aeolicus bound to NADH under anoxic conditions | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
7ZEH
 
 | | Human cytosolic 5' nucleotidase IIIB in complex with 3,4-diF-Bn7Guanine | | Descriptor: | 1,2-ETHANEDIOL, 7-methylguanosine phosphate-specific 5'-nucleotidase, D-ribulose, ... | | Authors: | Kubacka, D, Kozarski, M, Baranowski, M.R, Wojcik, R, Jemielity, J, Kowalska, J, Basquin, J. | | Deposit date: | 2022-03-31 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate-Based Design of Cytosolic Nucleotidase IIIB Inhibitors and Structural Insights into Inhibition Mechanism.
Pharmaceuticals, 15, 2022
|
|
5NIZ
 
 | | EthR complex | | Descriptor: | 1-[3-[5-[(1~{R},2~{R})-2-methylcyclopropyl]furan-2-yl]propanoyl]piperidine-4-carboxamide, HTH-type transcriptional regulator EthR, SULFATE ION | | Authors: | Pohl, E, Tatum, N.J, Cole, J.C, Baulard, A.R. | | Deposit date: | 2017-03-27 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New active leads for tuberculosis booster drugs by structure-based drug discovery.
Org. Biomol. Chem., 15, 2017
|
|
9BSN
 
 | |
9C09
 
 | | Structure of K2P13.1 (THIK1) S136P in lipid nanodisc | | Descriptor: | DECANE, DODECANE, HEXANE, ... | | Authors: | Roy-Chowdhury, S, Adberemane-Ali, F, Minor, D.L. | | Deposit date: | 2024-05-24 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Structure of the human K 2P 13.1 channel reveals a hydrophilic pore restriction and lipid cofactor site.
Nat.Struct.Mol.Biol., 32, 2025
|
|
7U6A
 
 | |
6G4Q
 
 | | Structure of human ADP-forming succinyl-CoA ligase complex SUCLG1-SUCLA2 | | Descriptor: | 1,2-ETHANEDIOL, Succinate--CoA ligase [ADP-forming] subunit beta, mitochondrial, ... | | Authors: | Bailey, H.J, Shrestha, L, Rembeza, E, Sorrell, F.J, Newman, J, Strain-Damerell, C, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2018-03-28 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of human ADP-forming succinyl-CoA ligase complex SUCLG1-SUCLA2
To Be Published
|
|
6S4H
 
 | | The crystal structure of glycogen phosphorylase in complex with 8 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(2-phenyl-1~{H}-imidazol-4-yl)oxane-3,4,5-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
