9GTG
 
 | | RIPK1 in complex with AZ"902 | | Descriptor: | (5R)-5-[(7-chloro-1H-indol-3-yl)methyl]-3-methylimidazolidine-2,4-dione, Receptor-interacting serine/threonine-protein kinase 1, ~{N}-[[(3~{S})-1-ethanoylpyrrolidin-3-yl]methyl]-~{N}-methyl-4-quinolin-7-yl-benzenesulfonamide | | Authors: | Petersen, J. | | Deposit date: | 2024-09-17 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery and Validation of a Novel Class of Necroptosis Inhibitors Targeting RIPK1.
Acs Chem.Biol., 20, 2025
|
|
7G0H
 
 | | Crystal Structure of human FABP4 in complex with (2R,3R)-2-(phenoxymethyl)-1-phenyl-pyrrolidine-3-carboxylic acid, i.e. SMILES C1C[C@H]([C@@H](N1c1ccccc1)COc1ccccc1)C(=O)O with IC50=0.168 microM | | Descriptor: | (2R,3S)-2-(phenoxymethyl)-1-phenylpyrrolidine-3-carboxylic acid, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6I4B
 
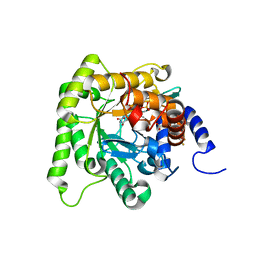 | | Plasmodium falciparum dihydroorotate dehydrogenase (DHODH) co-crystallized with 3-Hydroxy-1-methyl-5-((3-(trifluoromethyl)phenoxy)methyl)-1H-pyrazole-4-carboxylic acid | | Descriptor: | 1-methyl-3-oxidanyl-5-[[3-(trifluoromethyl)phenoxy]methyl]pyrazole-4-carboxylic acid, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Goyal, P, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S. | | Deposit date: | 2018-11-09 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Hydroxyazole scaffold-based Plasmodium falciparum dihydroorotate dehydrogenase inhibitors: Synthesis, biological evaluation and X-ray structural studies.
Eur J Med Chem, 163, 2018
|
|
5GQJ
 
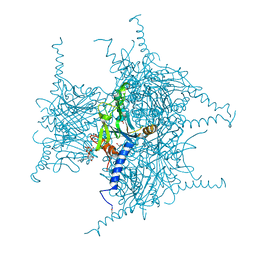 | | Crystal structure of Cypovirus Polyhedra mutant with deletion of Ser193 and Ala194 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
7FZ3
 
 | | Crystal Structure of human FABP4 in complex with 4-(4-chlorophenoxy)benzenesulfinic acid:sodium hydride, i.e. SMILES c1(Oc2ccc(cc2)Cl)ccc(cc1)[S@@](=O)O with IC50=7.4 microM | | Descriptor: | 1-[bis(oxidanyl)-$l^{3}-sulfanyl]-4-(4-chloranylphenoxy)benzene, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6RD9
 
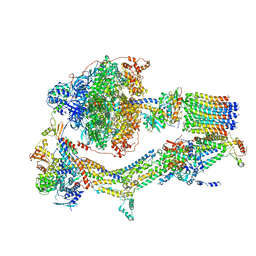 | | CryoEM structure of Polytomella F-ATP synthase, Primary rotary state 1, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
8SSS
 
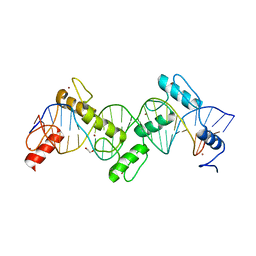 | | ZnFs 1-7 of CCCTC-binding factor (CTCF) Complexed with 23mer | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand (23mer) I, DNA Strand (23mer) II, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
6RBS
 
 | |
5N1T
 
 | | Crystal structure of complex between flavocytochrome c and copper chaperone CopC from T. paradoxus | | Descriptor: | COPPER (II) ION, CopC, Cytochrome C, ... | | Authors: | Osipov, E.M, Lilina, A.V, Tikhonova, T.V, Tsallagov, S.I, Popov, V.O. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-28 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the flavocytochrome c sulfide dehydrogenase associated with the copper-binding protein CopC from the haloalkaliphilic sulfur-oxidizing bacterium Thioalkalivibrio paradoxusARh 1.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6RBZ
 
 | |
6V3Q
 
 | | Crystal Structure of the Metallo-beta-Lactamase FIM-1 from Pseudomonas aeruginosa in the Mono-Zinc Form | | Descriptor: | ISOPROPYL ALCOHOL, Metallo-beta-lactamase FIM-1, ZINC ION | | Authors: | Kim, Y, Hatzos-Skintges, C, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-11-26 | | Release date: | 2020-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Metallo-beta-Lactamase FIM-1 from Pseudomonas aeruginosa in the Mono-Zinc Form
To Be Published
|
|
9GQ6
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog p5(1,2)-H2 | | Descriptor: | (2~{S},9~{S})-2-cyclohexyl-19,22-dimethoxy-11,17-dioxa-4-azatricyclo[16.2.2.0^{4,9}]docosa-1(20),18,21-triene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8THW
 
 | | Cac1 PIP motif bound to PCNA | | Descriptor: | Proliferating cell nuclear antigen,Chromatin assembly factor 1 subunit p90 | | Authors: | Veltri, E, Hoitsma, N.M, Dieckman, L. | | Deposit date: | 2023-07-18 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Interaction Between Yeast Chromatin Assembly Factor 1 and Proliferating Cell Nuclear Antigen.
J.Mol.Biol., 436, 2024
|
|
9GQ7
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog p5(2,1)-(E) | | Descriptor: | (2~{S},9~{S},14~{E})-2-cyclohexyl-19,22-dimethoxy-11,17-dioxa-4-azatricyclo[16.2.2.0^{4,9}]docosa-1(20),14,18,21-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GQ4
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog p5(3,1)-(E) | | Descriptor: | (2~{S},9~{S},16~{E})-2-cyclohexyl-21,24-dimethoxy-11,14,19-trioxa-4-azatricyclo[18.2.2.0^{4,9}]tetracosa-1(23),16,20(24),21-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GQB
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog p5(2,1)-Diol-II | | Descriptor: | (2~{S},9~{S},14~{S},15~{S})-2-cyclohexyl-19,22-dimethoxy-14,15-bis(oxidanyl)-11,17-dioxa-4-azatricyclo[16.2.2.0^{4,9}]docosa-1(21),18(22),19-triene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
6R71
 
 | | Crystal structure of human carbonic anhydrase isozyme XII with 2-(benzenesulfonyl)-4-chloro-N-(2-hydroxyethyl)-5-sulfamoyl-benzamide | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-~{N}-(2-hydroxyethyl)-2-(phenylsulfonyl)-5-sulfamoyl-benzamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2019-03-28 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Halogenated and di-substituted benzenesulfonamides as selective inhibitors of carbonic anhydrase isoforms.
Eur.J.Med.Chem., 185, 2020
|
|
7SS6
 
 | | Structure of Klebsiella LpxH in complex with JH-LPH-45 | | Descriptor: | 1,2-ETHANEDIOL, 5-{4-[3-chloro-5-(trifluoromethyl)phenyl]piperazine-1-sulfonyl}-N-[5-(hydroxyamino)-5-oxopentyl]-2,3-dihydro-1H-indole-1-carboxamide, MANGANESE (II) ION, ... | | Authors: | Cho, J, Cochrane, C.S, Zhou, P. | | Deposit date: | 2021-11-09 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Development of LpxH Inhibitors Chelating the Active Site Dimanganese Metal Cluster of LpxH.
Chemmedchem, 18, 2023
|
|
4YWI
 
 | | F96S/L167V Double mutant of Plasmodium Falciparum Triosephosphate Isomerase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Triosephosphate isomerase | | Authors: | Pareek, V, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2015-03-20 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Connecting Active-Site Loop Conformations and Catalysis in Triosephosphate Isomerase: Insights from a Rare Variation at Residue 96 in the Plasmodial Enzyme
Chembiochem, 17, 2016
|
|
6ZPV
 
 | | Structure of Unliganded MgGH51 a-L-Arabinofuranosidase Crystal Type 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6OT1
 
 | |
8U49
 
 | | The Apo Crystal Structure of BlCel9A from Glycoside Hydrolase Family 9 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Araujo, E.A, Polikarpov, I. | | Deposit date: | 2023-09-10 | | Release date: | 2024-02-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of cellulose depolymerization by the two-domain BlCel9A enzyme from the glycoside hydrolase family 9.
Carbohydr Polym, 329, 2024
|
|
8SGN
 
 | | Crystal structure of Epstein-Barr virus glycoprotein 350 (gp350) in complex with Cy651H02, a monoclonal antibody isolated from macaques immunized with a gp350 nanoparticle vaccine | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Joyce, M.G, Jensen, J.L, Chen, W.H, Kanekiyo, M. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-17 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for complement receptor engagement and virus neutralization through Epstein-Barr virus gp350.
Immunity, 58, 2025
|
|
9N3Q
 
 | |
9GOZ
 
 | | 4-Allyl syringol oxidase from Streptomyces cavernae: complex with eugenol | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-methoxy-4-(prop-2-en-1-yl)phenol, 4-allyl syringol oxidase from Streptomyces cavernae, ... | | Authors: | Mattevi, A, Alvigini, L. | | Deposit date: | 2024-09-06 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Kinetic and structural investigation of the 4-allyl syringol oxidase from Streptomyces cavernae.
Arch.Biochem.Biophys., 765, 2025
|
|
