2DLF
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 6.75 | | Descriptor: | PROTEIN (ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S) (HEAVY CHAIN)), PROTEIN (ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S)-KAPPA (LIGHT CHAIN)), SULFATE ION | | Authors: | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | Deposit date: | 1998-12-17 | | Release date: | 1999-12-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
2DSI
 
 | | Crystal structure of Glu171 to Arg mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Saraboji, K, Malathy Sony, S.M, Ponnuswamy, M.N, Kumarevel, T.S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-06-30 | | Release date: | 2006-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
2DTM
 
 | | Thermodynamic and structural analyses of hydrolytic mechanism by catalytic antibodies | | Descriptor: | IMMUNOGLOBULIN 6D9 | | Authors: | Oda, M, Ito, N, Tsumuraya, T, Suzuki, K, Fujii, I. | | Deposit date: | 2006-07-13 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
2DWC
 
 | | Crystal structure of Probable phosphoribosylglycinamide formyl transferase from Pyrococcus horikoshii OT3 complexed with ADP | | Descriptor: | 433aa long hypothetical phosphoribosylglycinamide formyl transferase, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Yoshikawa, S, Arai, R, Kamo-Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Probable phosphoribosylglycinamide formyl transferase from Pyrococcus horikoshii OT3 complexed with ADP
To be Published
|
|
3C36
 
 | |
2DVI
 
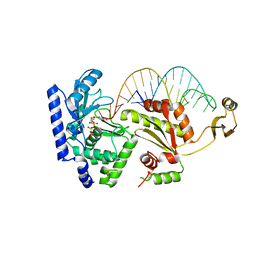 | | Complex structure of CCA-adding enzyme, mini-DCC and CTP | | Descriptor: | CCA-adding enzyme, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tomita, K, Ishitani, R, Fukai, S, Nureki, O. | | Deposit date: | 2006-07-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Complete crystallographic analysis of the dynamics of CCA sequence addition
Nature, 443, 2006
|
|
2EZ6
 
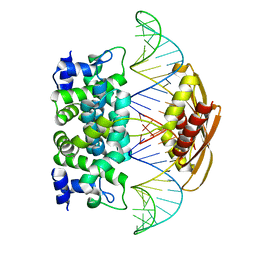 | | Crystal structure of Aquifex aeolicus RNase III (D44N) complexed with product of double-stranded RNA processing | | Descriptor: | 28-MER, MAGNESIUM ION, Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-11-10 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insight into the Mechanism of Double-Stranded RNA Processing by Ribonuclease III.
Cell(Cambridge,Mass.), 124, 2006
|
|
2ET2
 
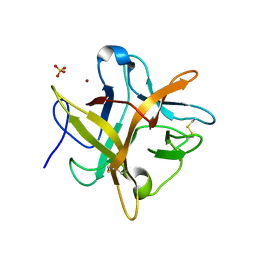 | | Crystal structure of an Asn to Ala mutant of Winged Bean Chymotrypsin Inhibitor protein | | Descriptor: | Chymotrypsin inhibitor 3, NICKEL (II) ION, SULFATE ION | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
3PFD
 
 | |
2EV5
 
 | | Bacillus subtilis manganese transport regulator (MNTR) bound to calcium | | Descriptor: | CALCIUM ION, Transcriptional regulator mntR | | Authors: | Kliegman, J.I, Griner, S.L, Helmann, J.D, Brennan, R.G, Glasfeld, A. | | Deposit date: | 2005-10-31 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Metal-Selective Activation of the Manganese Transport Regulator of Bacillus subtilis.
Biochemistry, 45, 2006
|
|
2F3T
 
 | |
2F1O
 
 | | Crystal Structure of NQO1 with Dicoumarol | | Descriptor: | BISHYDROXY[2H-1-BENZOPYRAN-2-ONE,1,2-BENZOPYRONE], FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Shaul, Y, Asher, G, Dym, O, Tsvetkov, P, Adler, J, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2005-11-15 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The crystal structure of NAD(P)H quinone oxidoreductase 1 in complex with its potent inhibitor dicoumarol.
Biochemistry, 45, 2006
|
|
2F7X
 
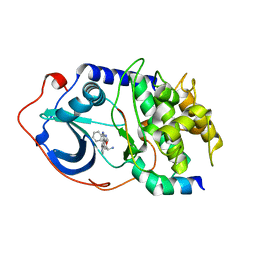 | | Protein Kinase A bound to (S)-2-(1H-Indol-3-yl)-1-[5-((E)-2-pyridin-4-yl-vinyl)-pyridin-3-yloxymethyl]-ethylamine | | Descriptor: | (1S)-2-(1H-INDOL-3-YL)-1-[({5-[(E)-2-PYRIDIN-4-YLVINYL]PYRIDIN-3-YL}OXY)METHYL]ETHYLAMINE, PKI, inhibitory peptide, ... | | Authors: | Li, Q, Li, T, Zhu, G.D, Gong, J, Claibone, A, Dalton, C, Luo, Y, Johnson, E.F, Shi, Y, Liu, X, Klinghofer, V, Bauch, J.L, Marsh, K.C, Bouska, J.J, Arries, S, De Jong, R, Oltersdorf, T, Stoll, V.S, Jakob, C.G, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2005-12-01 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of trans-3,4'-bispyridinylethylenes as potent and novel inhibitors of protein kinase B (PKB/Akt) for the treatment of cancer: Synthesis and biological evaluation.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2F5A
 
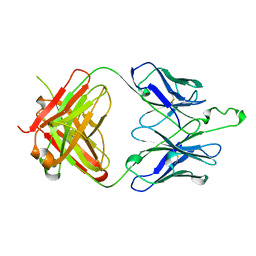 | | CRYSTAL STRUCTURE OF FAB' FROM THE HIV-1 NEUTRALIZING ANTIBODY 2F5 | | Descriptor: | PROTEIN (ANTIBODY 2F5 (HEAVY CHAIN)), PROTEIN (ANTIBODY 2F5 (LIGHT CHAIN)) | | Authors: | Bryson, S, Julien, J.P, Hynes, R.C, Pai, E.F. | | Deposit date: | 1999-04-08 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications.
J.Virol., 83, 2009
|
|
2F5N
 
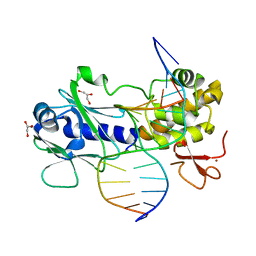 | | MutM crosslinked to undamaged DNA sampling A:T base pair IC1 | | Descriptor: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*C*GP*TP*CP*CP*AP*GP*GP*TP*CP*TP*AP*CP*C)-3', GLYCEROL, ... | | Authors: | Banerjee, A, Santos, W.L, Verdine, G.L. | | Deposit date: | 2005-11-26 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a DNA glycosylase searching for lesions.
Science, 311, 2006
|
|
2F60
 
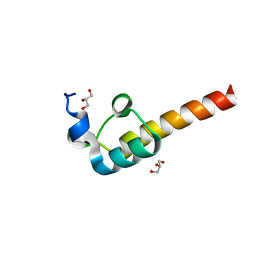 | | Crystal Structure of the Dihydrolipoamide Dehydrogenase (E3)-Binding Domain of Human E3-Binding Protein | | Descriptor: | GLYCEROL, Pyruvate dehydrogenase protein X component | | Authors: | Brautigam, C.A, Chuang, J.L, Wynn, R.M, Tomchick, D.R, Machius, M, Chuang, D.T. | | Deposit date: | 2005-11-28 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insight into Interactions between Dihydrolipoamide Dehydrogenase (E3) and E3 Binding Protein of Human Pyruvate Dehydrogenase Complex.
Structure, 14, 2006
|
|
2F9Y
 
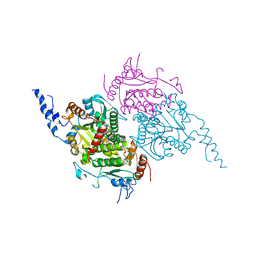 | | The Crystal Structure of The Carboxyltransferase Subunit of ACC from Escherichia coli | | Descriptor: | Acetyl-CoA carboxylase, Carboxyltransferase alpha chain, Acetyl-coenzyme A carboxylase carboxyl transferase subunit beta, ... | | Authors: | Bilder, P.W. | | Deposit date: | 2005-12-06 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Structure of the Carboxyltransferase Component of Acetyl-CoA Carboxylase Reveals a Zinc-Binding Motif Unique to the Bacterial Enzyme(,).
Biochemistry, 45, 2006
|
|
2FDA
 
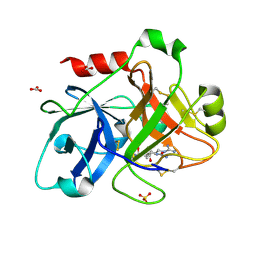 | | Crystal Structure of the Catalytic Domain of Human Coagulation Factor XIa in Complex with alpha-Ketothiazole Arginine Derived Ligand | | Descriptor: | BICARBONATE ION, Coagulation factor XI, N~2~-(AMINOCARBONYL)-N~1~-{4-{[AMINO(IMINO)METHYL]AMINO}-1-[HYDROXY(1,3-THIAZOL-2-YL)METHYL]BUTYL}VALINAMIDE, ... | | Authors: | Jin, L, Pandey, P, Babine, R.E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-12-13 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis, SAR exploration, and X-ray crystal structures of factor XIa inhibitors containing an alpha-ketothiazole arginine
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2F9T
 
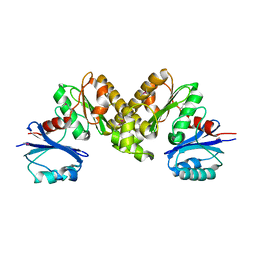 | | Structure of the type III CoaA from Pseudomonas aeruginosa | | Descriptor: | Pantothenate Kinase | | Authors: | Leonardi, R, Yun, M.K, Chohnan, S, White, S.W, Rock, C.O, Jackowski, S. | | Deposit date: | 2005-12-06 | | Release date: | 2006-08-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prokaryotic Type II and Type III Pantothenate Kinases: The Same Monomer Fold Creates Dimers with Distinct Catalytic Properties.
Structure, 14, 2006
|
|
2FGI
 
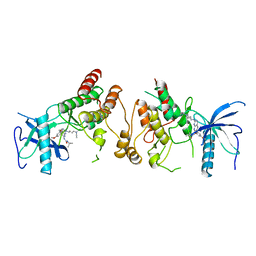 | | CRYSTAL STRUCTURE OF THE TYROSINE KINASE DOMAIN OF FGF RECEPTOR 1 IN COMPLEX WITH INHIBITOR PD173074 | | Descriptor: | 1-TERT-BUTYL-3-[6-(3,5-DIMETHOXY-PHENYL)-2-(4-DIETHYLAMINO-BUTYLAMINO)-PYRIDO[2,3-D]PYRIMIDIN-7-YL]-UREA, PROTEIN (FIBROBLAST GROWTH FACTOR (FGF) RECEPTOR 1) | | Authors: | Mohammadi, M, Froum, S, Hamby, J.M, Schroeder, M, Panek, R.L, Lu, G.H, Eliseenkova, A.V, Green, D, Schlessinger, J, Hubbard, S.R. | | Deposit date: | 1998-09-15 | | Release date: | 1999-09-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain.
EMBO J., 17, 1998
|
|
2FG7
 
 | | N-succinyl-L-ornithine transcarbamylase from B. fragilis complexed with carbamoyl phosphate and N-succinyl-L-norvaline | | Descriptor: | N-(3-CARBOXYPROPANOYL)-L-NORVALINE, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, SULFATE ION, ... | | Authors: | Shi, D, Yu, X, Malamy, M.H, Allewell, N.M, Tuchman, M. | | Deposit date: | 2005-12-21 | | Release date: | 2006-05-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and catalytic mechanism of a novel N-succinyl-L-ornithine transcarbamylase in arginine biosynthesis of Bacteroides fragilis.
J.Biol.Chem., 281, 2006
|
|
3C34
 
 | |
2FME
 
 | | Crystal structure of the mitotic kinesin eg5 (ksp) in complex with mg-adp and (r)-4-(3-hydroxyphenyl)-n,n,7,8-tetramethyl-3,4-dihydroisoquinoline-2(1h)-carboxamide | | Descriptor: | (4R)-4-(3-HYDROXYPHENYL)-N,N,7,8-TETRAMETHYL-3,4-DIHYDROISOQUINOLINE-2(1H)-CARBOXAMIDE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Sheriff, S. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of human mitotic kinesin Eg5: Characterization of the 4-phenyl-tetrahydroisoquinoline lead series
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2EVX
 
 | |
2EV6
 
 | | Bacillus subtilis manganese transport regulator (MNTR) bound to zinc | | Descriptor: | CITRATE ANION, GLYCEROL, Transcriptional regulator mntR, ... | | Authors: | Kliegman, J.I, Griner, S.L, Helmann, J.D, Brennan, R.G, Glasfeld, A. | | Deposit date: | 2005-10-31 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Metal-Selective Activation of the Manganese Transport Regulator of Bacillus subtilis.
Biochemistry, 45, 2006
|
|
