7CDN
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 42 kGy (9000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDP
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 42 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDQ
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 83 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDR
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 210 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDK
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 42 kGy (4500 images from 1st half of data set) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDO
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 21 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDU
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 1700 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDT
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 830 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDS
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 420 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
6Z74
 
 | |
6DWH
 
 | | Crystal structure of complex of BBKI and Bovine Trypsin | | Descriptor: | CHLORIDE ION, Cationic trypsin, Kunitz-type inihibitor, ... | | Authors: | Li, M, Wlodawer, A. | | Deposit date: | 2018-06-26 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the complex of a kallikrein inhibitor from Bauhinia bauhinioides with trypsin and modeling of kallikrein complexes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5KLO
 
 | | Crystal structure of thioacyl intermediate in 2-aminomuconate 6-semialdehyde dehydrogenase N169A | | Descriptor: | (2Z,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
5KLM
 
 | | Crystal structure of 2-hydroxymuconate-6-semialdehyde derived intermediate in NAD(+)-bound 2-aminomuconate 6-semialdehyde dehydrogenase N169D | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
6DMD
 
 | | ppGpp Riboswitch bound to ppGpp, manganese chloride structure | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Peselis, A, Serganov, A. | | Deposit date: | 2018-06-05 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | ykkC riboswitches employ an add-on helix to adjust specificity for polyanionic ligands.
Nat. Chem. Biol., 14, 2018
|
|
8RZH
 
 | | ZgGH129 from Zobellia galactanivorans in complex with the inhibitor AD-DGJ (3,6-anhydro-D-1-deoxygalactonojirimycin). | | Descriptor: | (1~{R},4~{S},5~{R},8~{S})-6-oxa-2-azabicyclo[3.2.1]octane-4,8-diol, 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8RZJ
 
 | | ZgGH129 from Zobellia galactanivorans in complex with the inhibitor ADG-IF (3,6-anhydro-D-galacto-isofagomine). | | Descriptor: | (1~{R},5~{R},8~{S})-6-oxa-3-azabicyclo[3.2.1]octan-8-ol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8RZI
 
 | | ZgGH129 from Zobellia galactanivorans soaked with 1,2-diF-ADG (3,6-Anhydro-2-deoxy-2-fluoro-a-D-galactopyranosyl fluoride) resulting in a trapped glycosyl-enzyme intermediate. | | Descriptor: | (1~{R},4~{S},5~{S},8~{S})-4-fluoranyl-2,6-dioxabicyclo[3.2.1]octan-8-ol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 2024
|
|
5KLN
 
 | | Crystal structure of 2-aminomuconate 6-semialdehyde dehydrogenase N169A in complex with NAD+ | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
8RZG
 
 | | ZgGH129 from Zobellia galactanivorans soaked with the product of the reaction ADG (3,6-anhydro-D-galactose). | | Descriptor: | (1~{R},4~{S},5~{R},8~{S})-2,6-dioxabicyclo[3.2.1]octane-4,8-diol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 2024
|
|
7D0Y
 
 | |
4PA7
 
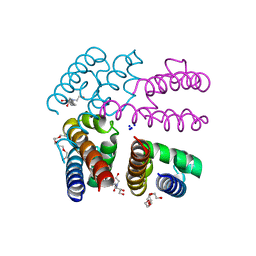 | | Structure of NavMS pore and C-terminal domain crystallised in presence of channel blocking compound | | Descriptor: | DODECAETHYLENE GLYCOL, HEGA-10, Ion transport protein, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5O8N
 
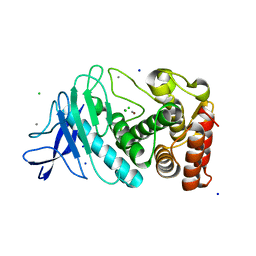 | | Structure of thermolysin at room temperature via a method of acoustically induced rotation. | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Axford, D.N, Burton, C, Docker, P, Prince, M, Topham, P.D. | | Deposit date: | 2017-06-13 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An acoustic on-chip goniometer for room temperature macromolecular crystallography.
Lab Chip, 17, 2017
|
|
5KWE
 
 | |
4NLP
 
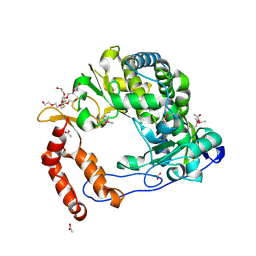 | | Poliovirus Polymerase - C290V Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
5NJC
 
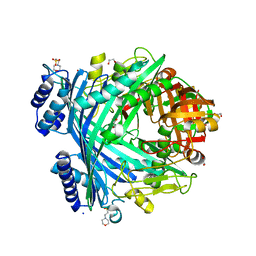 | | E. coli Microcin-processing metalloprotease TldD/E (TldD E263A mutant) with hexapeptide bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Metalloprotease PmbA, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
