7T1I
 
 | | Crystal structure of CAB1 Pantothenate Kinase from Saccharomyces cerevisiae in complex with compound YU385597 | | Descriptor: | (8S)-2-{[(4-tert-butylphenyl)methyl]amino}-5-[(piperidin-1-yl)methyl][1,2,4]triazolo[1,5-a]pyrimidin-7(6H)-one, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gihaz, S, Ben Mamoun, C. | | Deposit date: | 2021-12-02 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-resolution crystal structure and chemical screening reveal pantothenate kinase as a new target for antifungal development.
Structure, 30, 2022
|
|
8ACS
 
 | | Crystal structure of FMO from Janthinobacterium svalbardensis | | Descriptor: | DI(HYDROXYETHYL)ETHER, FAD-dependent oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Polidori, N, Galuska, P, Gruber, K. | | Deposit date: | 2022-07-06 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Cold-Active Flavin-Dependent Monooxygenase from Janthinobacterium svalbardensis Unlocks Applications of Baeyer-Villiger Monooxygenases at Low Temperature.
Acs Catalysis, 13, 2023
|
|
7AYC
 
 | | Crystal Structure of human mitochondrial 2-Enoyl Thioester Reductase (MECR) with single mutation G165Q | | Descriptor: | CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase, mitochondrial | | Authors: | Rahman, M.T, Koski, M.K, Autio, K.J, Kastaniotis, A.J, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2020-11-12 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | An engineered variant of MECR reductase reveals indispensability of long-chain acyl-ACPs for mitochondrial respiration.
Nat Commun, 14, 2023
|
|
6O94
 
 | | Structure of the IRAK4 kinase domain with compound 17 | | Descriptor: | CALCIUM ION, Interleukin-1 receptor-associated kinase 4, N-{5-[4-(hydroxymethyl)piperidin-1-yl]-1-methyl-2-(morpholin-4-yl)-1H-benzimidazol-6-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Authors: | Yu, C, Drobnick, J, Bryan, M.C, Kiefer, J, Lupardus, P.J. | | Deposit date: | 2019-03-13 | | Release date: | 2019-05-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Development of Potent and Selective Pyrazolopyrimidine IRAK4 Inhibitors.
J.Med.Chem., 62, 2019
|
|
7T1H
 
 | | Crystal structure of CAB1 Pantothenate Kinase from Saccharomyces cerevisiae in complex with compound YU281445 | | Descriptor: | (8R)-2-{[(4-tert-butylphenyl)methyl]amino}-5-[(morpholin-4-yl)methyl][1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, GLYCEROL, Pantothenate kinase CAB1 | | Authors: | Gihaz, S, Ben Mamoun, C. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structure and chemical screening reveal pantothenate kinase as a new target for antifungal development.
Structure, 30, 2022
|
|
7AYZ
 
 | |
6BZF
 
 | | Structure of S. cerevisiae Zip2:Spo16 complex, C2 form | | Descriptor: | Protein ZIP2, Sporulation-specific protein 16 | | Authors: | Arora, K, Corbett, K.D. | | Deposit date: | 2017-12-23 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.286 Å) | | Cite: | The conserved XPF:ERCC1-like Zip2:Spo16 complex controls meiotic crossover formation through structure-specific DNA binding.
Nucleic Acids Res., 47, 2019
|
|
7AVN
 
 | | Structure of marine actinobacteria clade rhodopsin (MacR) in orange form in P1 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, ... | | Authors: | Gushchin, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gordeliy, V. | | Deposit date: | 2020-11-05 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Alternating Access Mechanism in a Minimalistic Light-Driven Proton Pump
To Be Published
|
|
6XH0
 
 | |
6XHH
 
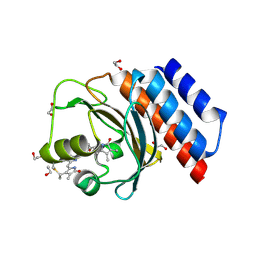 | | Far-red absorbing dark state of JSC1_58120g3 with bound 18-1, 18-2 dihydrobiliverdin IXa (DHBV), the native chromophore precursor | | Descriptor: | 1,2-ETHANEDIOL, JSC1_58120g3, mesobiliverdin IX(alpha) | | Authors: | Moreno, M.V, Rockwell, N.C, Fisher, A.J, Lagarias, J.C. | | Deposit date: | 2020-06-18 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A far-red cyanobacteriochrome lineage specific for verdins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6C08
 
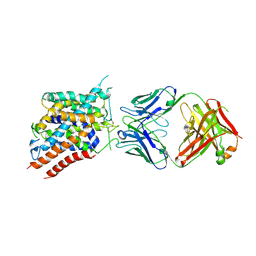 | |
7AZ0
 
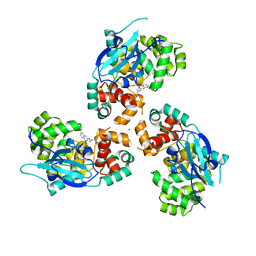 | |
7SMZ
 
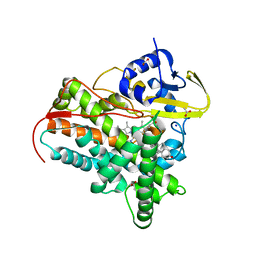 | | X-ray crystal structure of CYP142A3 from Mycobacterium Marinum in complex with 4-cholesten-3-one | | Descriptor: | (8ALPHA,9BETA)-CHOLEST-4-EN-3-ONE, ACETATE ION, Cytochrome P450 142A3, ... | | Authors: | Ghith, A, Bruning, J.B, Bell, S.G. | | Deposit date: | 2021-10-27 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Structures of the Steroid Binding CYP142 Cytochrome P450 Enzymes from Mycobacterium ulcerans and Mycobacterium marinum.
Acs Infect Dis., 8, 2022
|
|
6C18
 
 | | FGFR1 kinase complex with inhibitor SN37115 | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-{(3S)-1-[(2E)-4-(dimethylamino)but-2-enoyl]pyrrolidin-3-yl}-7-[(propan-2-yl)amino]-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Yosaatmadja, Y, Smaill, J.B, Squire, C.J. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Understanding the structural requirements for covalent inhibition of FGFR1-3
To Be Published
|
|
8AJP
 
 | |
7AYB
 
 | | Crystal Structure of wild type human mitochondrial 2-Enoyl Thioester Reductase (MECR) | | Descriptor: | ACETIC ACID, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase, ... | | Authors: | Rahman, M.T, Koski, M.K, Autio, K.J, Kastaniotis, A.J, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2020-11-12 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An engineered variant of MECR reductase reveals indispensability of long-chain acyl-ACPs for mitochondrial respiration.
Nat Commun, 14, 2023
|
|
6OIU
 
 | |
8AHU
 
 | | Crystal structure of D-amino acid aminotrensferase from Haliscomenobacter hydrossis complexed with D-cycloserine | | Descriptor: | Aminotransferase class IV, GLYCEROL, [5-hydroxy-6-methyl-4-({[(4E)-3-oxo-1,2-oxazolidin-4-ylidene]amino}methyl)pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Bakunova, A.K, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-22 | | Release date: | 2022-08-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Mechanism of D-Cycloserine Inhibition of D-Amino Acid Transaminase from Haliscomenobacter hydrossis.
Biochemistry Mosc., 88, 2023
|
|
7AVO
 
 | | Structure of marine actinobacteria clade rhodopsin (MacR) in orange form in P1211 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, ... | | Authors: | Gushchin, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gordeliy, V. | | Deposit date: | 2020-11-05 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Alternating Access Mechanism in a Minimalistic Light-Driven Proton Pump
To Be Published
|
|
8SHK
 
 | |
8AG2
 
 | | Crystal structure of the BPTF bromodomain in complex with BI-7190 | | Descriptor: | 5-[3-methoxy-4-[1-(4-methylpiperazin-1-yl)cyclopropyl]phenyl]-1,3,4-trimethyl-pyridin-2-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Bader, G, Boettcher, J, Wolkerstorfer, B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.025 Å) | | Cite: | Discovery of a Chemical Probe to Study Implications of BPTF Bromodomain Inhibition in Cellular and in vivo Experiments.
Chemmedchem, 18, 2023
|
|
8AQD
 
 | | Hydrophobic probe bound to Streptavidin - 1 | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[2-[6-(dimethylamino)-1,3-bis(oxidanylidene)benzo[de]isoquinolin-2-yl]ethyl]pentanamide, Streptavidin | | Authors: | Igareta, N.V, Ward, T.R. | | Deposit date: | 2022-08-12 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Hydrophobic probe bound to Streptavidin - 1
To Be Published
|
|
7B1T
 
 | | Crystal structure of BRD4(1) in complex with the inhibitor MPM6 | | Descriptor: | 3-(5-azanyl-2-chloranyl-phenyl)-1-methyl-4,7-dihydro-2~{H}-cyclohepta[c]pyrrol-8-one, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE | | Authors: | Huegle, M. | | Deposit date: | 2020-11-25 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A novel pan-selective bromodomain inhibitor for epigenetic drug design.
Eur.J.Med.Chem., 249, 2023
|
|
6ZAX
 
 | | Nitrite-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) at low dose (0.5 MGy) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
6GFK
 
 | | delta-N METTL16 MTase domain | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, U6 small nuclear RNA (adenine-(43)-N(6))-methyltransferase | | Authors: | Chen, K.M, Mendel, M, Homolka, D, McCarthy, A.A, Pillai, R.S. | | Deposit date: | 2018-04-30 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Methylation of Structured RNA by the m6A Writer METTL16 Is Essential for Mouse Embryonic Development.
Mol. Cell, 71, 2018
|
|
