3M7A
 
 | |
3NQK
 
 | |
4HM9
 
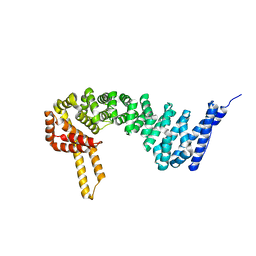 | | Crystal structure of full-length human catenin-beta-like 1 | | Descriptor: | Beta-catenin-like protein 1 | | Authors: | Du, Z, Huang, X, Wang, G, Wu, Y. | | Deposit date: | 2012-10-18 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1001 Å) | | Cite: | The structure of full-length human CTNNBL1 reveals a distinct member of the armadillo-repeat protein family.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4IB2
 
 | |
3NQO
 
 | |
3NRF
 
 | |
4I8S
 
 | |
4IAG
 
 | | Crystal structure of ZbmA, the zorbamycin binding protein from Streptomyces flavoviridis | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Zbm binding protein | | Authors: | Cuff, M.E, Bigelow, L, Bruno, C.J.P, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-12-06 | | Release date: | 2013-02-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Zorbamycin-Binding Protein ZbmA, the Primary Self-Resistance Element in Streptomyces flavoviridis ATCC21892.
Biochemistry, 54, 2015
|
|
3O0R
 
 | | Crystal structure of nitric oxide reductase from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | CALCIUM ION, FE (III) ION, HEME C, ... | | Authors: | Hino, T, Matsumoto, Y, Nagano, S, Sugimoto, H, Fukumori, Y, Murata, T, Iwata, S, Shiro, Y. | | Deposit date: | 2010-07-20 | | Release date: | 2010-12-29 | | Last modified: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of biological N2O generation by bacterial nitric oxide reductase
Science, 330, 2010
|
|
4HPJ
 
 | | Crystal structure of Tryptophan Synthase at 1.45 A resolution in complex with 2-aminophenol quinonoid in the beta site and the F9 inhibitor in the alpha site | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-3-[(2-hydroxyphenyl)amino]propanoic acid, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, BICINE, ... | | Authors: | Hilario, E, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2012-10-23 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Allostery and substrate channeling in the tryptophan synthase bienzyme complex: evidence for two subunit conformations and four quaternary states.
Biochemistry, 52, 2013
|
|
4IAW
 
 | | Engineered human lipocalin 2 (C26) in complex with Y-DTPA | | Descriptor: | N-{(1S,2S)-2-[bis(carboxymethyl)amino]cyclohexyl}-N-{(2R)-2-[bis(carboxymethyl)amino]-3-[4-({[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]carbamothioyl}amino)phenyl]propyl}glycine, Neutrophil gelatinase-associated lipocalin, YTTRIUM (III) ION | | Authors: | Eichinger, A, Skerra, A. | | Deposit date: | 2012-12-07 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-guided engineering of Anticalins with improved binding behavior and biochemical characteristics for application in radio-immuno imaging and/or therapy
J.Struct.Biol., 185, 2014
|
|
4I5Z
 
 | |
4I5Y
 
 | |
4I6T
 
 | |
4IK6
 
 | |
7UZN
 
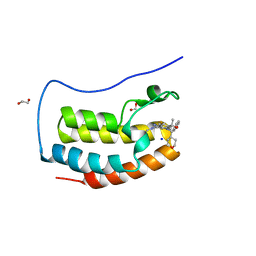 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH BMT-206059 AKA 2-{(3M)-3-(1,4-DIMETHYL-1H-1,2,3-TRIAZOL-5-YL)-8-FLUORO-5-[(S)-(OXAN-4-YL)(PHENYL)METHYL]-5H-PYRIDO[3,2-b]INDOL-7-YL}PROPAN-2-OL, TRIPLY DEUTERATED ON THE 4-METHYL GROUP | | Descriptor: | 1,2-ETHANEDIOL, 2-{(3M)-3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-8-fluoro-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol, Bromodomain-containing protein 4, ... | | Authors: | Sheriff, S. | | Deposit date: | 2022-05-09 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.685 Å) | | Cite: | Development of BET Inhibitors as Potential Treatments for Cancer: Optimization of Pharmacokinetic Properties.
Acs Med.Chem.Lett., 13, 2022
|
|
4IGG
 
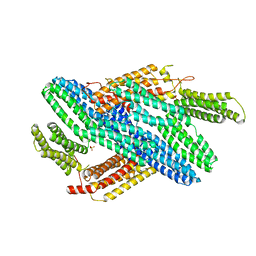 | |
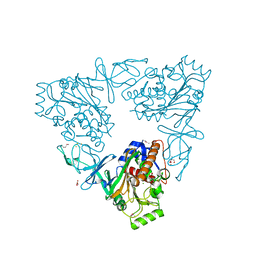
3N0Q
 
 | |
3N6W
 
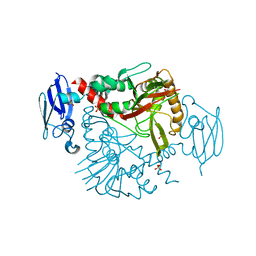 | | Crystal structure of human gamma-butyrobetaine hydroxylase | | Descriptor: | Gamma-butyrobetaine dioxygenase, SULFATE ION, ZINC ION | | Authors: | Rumnieks, J, Zeltins, A, Leonchiks, A, Kazaks, A, Kotelovica, S, Tars, K. | | Deposit date: | 2010-05-26 | | Release date: | 2010-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human gamma-butyrobetaine hydroxylase.
Biochem.Biophys.Res.Commun., 398, 2010
|
|
3MWX
 
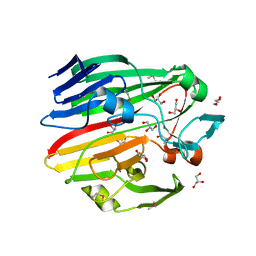 | |
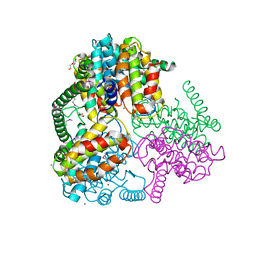
3MZO
 
 | |
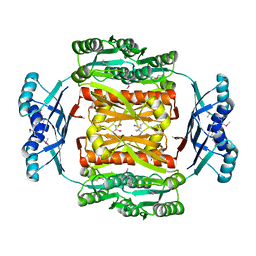
3N0V
 
 | |
3MW8
 
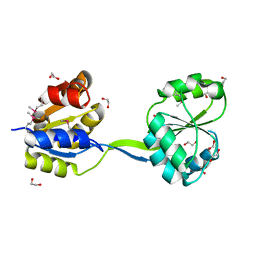 | |
4GNE
 
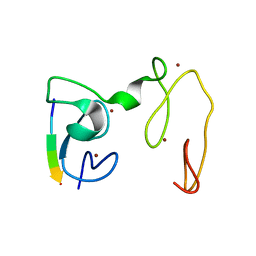 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3 peptide 1-7 | | Descriptor: | Histone H3.3, Histone-lysine N-methyltransferase NSD3, ZINC ION | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
3MYV
 
 | |
