8V4M
 
 | | CCP5 in complex with microtubules class3 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-29 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation
Nature, 2024
|
|
8V4L
 
 | | CCP5 in complex with microtubules class2 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-29 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation
Nature, 2024
|
|
8V4K
 
 | | CCP5 in complex with microtubules class1 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-29 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation
Nature, 2024
|
|
8V4J
 
 | | Phosphoheptose isomerase GMHA from Burkholderia pseudomallei bound to inhibitor Mut148233 | | Descriptor: | 1-deoxy-1-[formyl(hydroxy)amino]-5-O-phosphono-D-ribitol, CHLORIDE ION, Phosphoheptose isomerase, ... | | Authors: | Junop, M.S, Brown, C, Szabla, R. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Potentiating Activity of GmhA Inhibitors on Gram-Negative Bacteria.
J.Med.Chem., 67, 2024
|
|
8V4H
 
 | | X-ray structure of the NADP-dependent reductase from Campylobacter jejuni responsible for the synthesis of CDP-glucitol in the presence of CDP-glucitol | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Putative nucleotide sugar dehydratase, ... | | Authors: | Thoden, J.B, Schumann, M.E, Holden, H.M, Raushel, F.M. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biosynthesis of Cytidine Diphosphate-6-d-Glucitol for the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 63, 2024
|
|
8V4G
 
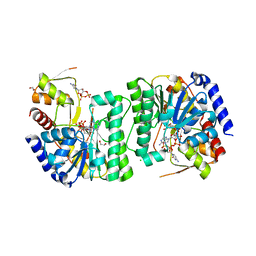 | | X-ray structure of the NADP-dependent reductase from Campylobacter jejuni responsible for the synthesis of CDP-glucitol in the presence of CDP and NADP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CYTIDINE-5'-DIPHOSPHATE, ... | | Authors: | Schumann, M.E, Thoden, J.B, Holden, H.M, Raushel, F.M. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biosynthesis of Cytidine Diphosphate-6-d-Glucitol for the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 63, 2024
|
|
8V4F
 
 | |
8V4A
 
 | | Proteus vulgaris tryptophan indole-lyase complexed with L-ethionine | | Descriptor: | (2E)-4-(ethylsulfanyl)-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}butanoic acid, (E)-S-ethyl-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-homocysteine, DIMETHYL SULFOXIDE, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2023-11-28 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Proteus vulgaris tryptophan indole-lyase complexed with L-ethionine
To Be Published
|
|
8V49
 
 | |
8V48
 
 | |
8V47
 
 | |
8V46
 
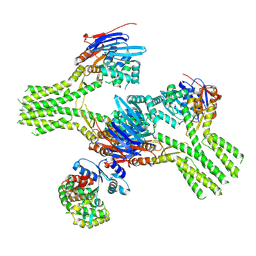 | |
8V45
 
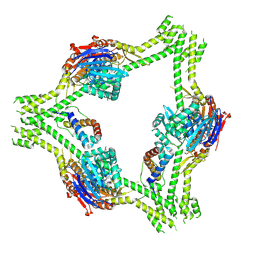 | |
8V44
 
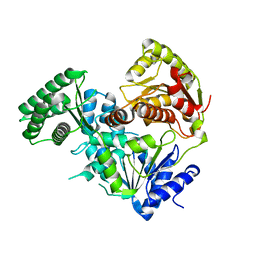 | |
8V3S
 
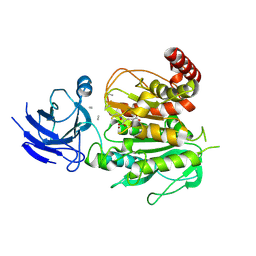 | | Structure of CCP5 class3 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation
Nature, 2024
|
|
8V3R
 
 | | Structure of CCP5 class2 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation
Nature, 2024
|
|
8V3Q
 
 | | Structure of CCP5 class1 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation
Nature, 2024
|
|
8V3P
 
 | | CCP5 in complex with Glu-P-peptide 2 transition state analog | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, Tubulin beta-2A chain, ZINC ION | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation
Nature, 2024
|
|
8V3O
 
 | | CCP5 in complex with Glu-P-peptide 1 transition state analog | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, D-MALATE, POTASSIUM ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation
Nature, 2024
|
|
8V3N
 
 | | CCP5 in complex with Glu-P-Glu transition state analog | | Descriptor: | (2S)-2-{[(S)-[(3S)-3-acetamido-4-(ethylamino)-4-oxobutyl](hydroxy)phosphoryl]methyl}pentanedioic acid, Cytosolic carboxypeptidase-like protein 5, D-MALATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation
Nature, 2024
|
|
8V3M
 
 | | CCP5 apo structure | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, D-MALATE, IMIDAZOLE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation
Nature, 2024
|
|
8V3L
 
 | |
8V3K
 
 | | F95S-F198S epi-Isozizaene Synthase: complex with 3 Mg2+, inorganic pyrophosphate, and benzyl triethyl ammonium cation | | Descriptor: | DIPHOSPHATE, Epi-isozizaene synthase, GLYCEROL, ... | | Authors: | Christianson, D.W, Eaton, S.A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Based Engineering of a Sesquiterpene Cyclase to Generate an Alcohol Product: Conversion of epi -Isozizaene Synthase into alpha-Bisabolol Synthase.
Biochemistry, 63, 2024
|
|
8V3D
 
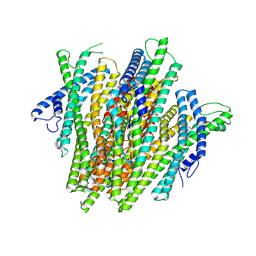 | | AgamOR28 structure bound to 2,4,5-trimethylthiazole | | Descriptor: | 2,4,5-trimethyl-1,3-thiazole, OR28, Odorant receptor Orco | | Authors: | Zhao, J, del Marmol, J. | | Deposit date: | 2023-11-27 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of odor sensing by insect heteromeric odorant receptors.
Science, 384, 2024
|
|
8V3C
 
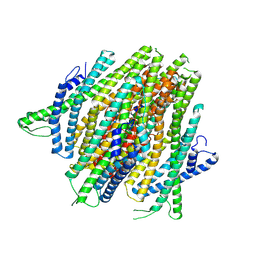 | | AgamOR28 structure without ligand | | Descriptor: | OR28, Odorant receptor Orco | | Authors: | Zhao, J, del Marmol, J. | | Deposit date: | 2023-11-27 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structural basis of odor sensing by insect heteromeric odorant receptors.
Science, 384, 2024
|
|
