6C31
 
 | | Crystal structure of TetR family protein Rv0078 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*TP*TP*AP*CP*CP*GP*GP*CP*AP*GP*TP*CP*TP*GP*CP*TP*TP*GP*TP*AP*AP*A)-3'), DNA (5'-D(P*AP*CP*AP*AP*GP*CP*AP*GP*AP*CP*TP*GP*CP*CP*GP*GP*TP*AP*AP*C)-3'), TetR family transcriptional regulator | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2018-01-09 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cytokinin Signaling in Mycobacterium tuberculosis.
MBio, 9, 2018
|
|
6BGN
 
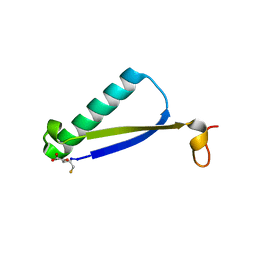 | | Crystal Structure of 4-Oxalocrotonate Tautomerase After Incubation with 5-Fluoro-2-hydroxy-2,4-pentadienoate | | Descriptor: | 2-hydroxymuconate tautomerase, 5-fluoranyl-2-oxidanylidene-pentanoic acid, GLYCEROL, ... | | Authors: | Zhang, Y, Li, W, Stack, T. | | Deposit date: | 2017-10-29 | | Release date: | 2018-02-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Inactivation of 4-Oxalocrotonate Tautomerase by 5-Halo-2-hydroxy-2,4-pentadienoates.
Biochemistry, 57, 2018
|
|
1F69
 
 | |
1M5U
 
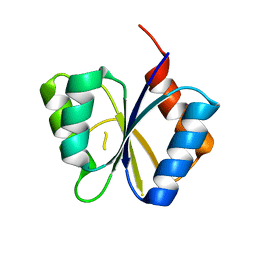 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK. STRUCTURE AT PH 8.0 IN THE APO-FORM | | Descriptor: | cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystallographic and biochemical studies of DivK reveal novel features of an essential response regulator in Caulobacter crescentus
J.Biol.Chem., 277, 2002
|
|
6BPO
 
 | |
7G00
 
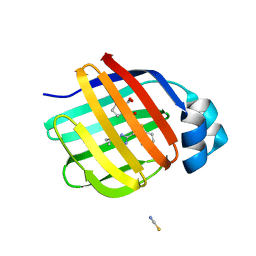 | | Crystal Structure of human FABP1 in complex with 2-[[3-(5-tert-butyl-1,2,4-oxadiazol-3-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl]cyclopentene-1-carboxylic acid | | Descriptor: | 2-{[(3P)-3-(5-tert-butyl-1,2,4-oxadiazol-3-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl}cyclopent-1-ene-1-carboxylic acid, Fatty acid-binding protein, liver, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Neidhart, W, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a human FABP1 complex
To be published
|
|
1MCF
 
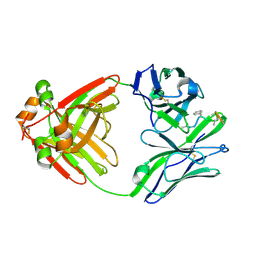 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | Immunoglobulin lambda-1 light chain, PEPTIDE N-ACETYL-L-GLN-D-PHE-L-HIS-D-PRO-B-ALA-B-ALA-OH | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2022-04-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1M8Q
 
 | | Molecular Models of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle | | Descriptor: | Skeletal muscle Actin, Skeletal muscle Myosin II, Skeletal muscle Myosin II Essential Light Chain, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-07-25 | | Release date: | 2002-09-10 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular modeling of averaged rigor crossbridges from tomograms of insect flight muscle.
J.Struct.Biol., 138, 2002
|
|
7G81
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z104474512 | | Descriptor: | 1-(2-ethoxyphenyl)piperazine, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
1F9I
 
 | | CRYSTAL STRUCTURE OF THE PHOTOACTIVE YELLOW PROTEIN MUTANT Y42F | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Brudler, R, Meyer, T.E, Genick, U.K, Tollin, G, Getzoff, E.D. | | Deposit date: | 2000-07-10 | | Release date: | 2000-07-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Coupling of hydrogen bonding to chromophore conformation and function in photoactive yellow protein.
Biochemistry, 39, 2000
|
|
7G9J
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with PCM-0102281-001 | | Descriptor: | 1-(3,4-dihydropyrazin-1(2H)-yl)ethan-1-one, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
6BX4
 
 | |
1F52
 
 | | CRYSTAL STRUCTURE OF GLUTAMINE SYNTHETASE FROM SALMONELLA TYPHIMURIUM CO-CRYSTALLIZED WITH ADP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLUTAMINE SYNTHETASE, ... | | Authors: | Gill, H.S, Pfluegl, G.M.U, Eisenberg, D. | | Deposit date: | 2000-06-12 | | Release date: | 2000-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The crystal structure of phosphinothricin in the active site of glutamine synthetase illuminates the mechanism of enzymatic inhibition.
Biochemistry, 40, 2001
|
|
7GJX
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-3ccb8ef6-1 (Mpro-P0744) | | Descriptor: | (4S)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
6BXQ
 
 | | Crystal Structure of HLA-B*57:01 with an HIV peptide RKV | | Descriptor: | Beta-2-microglobulin, HIV peptide RKV, MHC class I antigen | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-12-18 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification of Native and Posttranslationally Modified HLA-B*57:01-Restricted HIV Envelope Derived Epitopes Using Immunoproteomics.
Proteomics, 18, 2018
|
|
6BYP
 
 | | Structure of PL24 family Polysaccharide lyase-LOR107 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ulaganathan, T.S, Cygler, M. | | Deposit date: | 2017-12-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function analyses of a PL24 family ulvan lyase reveal key features and suggest its catalytic mechanism.
J. Biol. Chem., 293, 2018
|
|
1MHZ
 
 | | METHANE MONOOXYGENASE HYDROXYLASE | | Descriptor: | FE (III) ION, METHANE MONOOXYGENASE HYDROXYLASE | | Authors: | Elango, N, Radhakrishnan, R, Froland, W.A, Waller, B.J, Earhart, C.A, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1996-10-21 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b
Protein Sci., 6, 1997
|
|
1F6T
 
 | | STRUCTURE OF THE NUCLEOSIDE DIPHOSPHATE KINASE/ALPHA-BORANO(RP)-TDP.MG COMPLEX | | Descriptor: | 2*-DEOXY-THYMIDINE-5*-ALPHA BORANO DIPHOSPHATE (ISOMER RP), MAGNESIUM ION, PROTEIN (NUCLEOSIDE DIPHOSPHATE KINASE) | | Authors: | Guerreiro, C, Boretto, J, Janin, J, Veron, M, Canard, B, Schneider, B, Sarfati, S, Deville-Bonne, D. | | Deposit date: | 2000-06-23 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for activation of alpha-boranophosphate nucleotide analogues targeting drug-resistant reverse transcriptase.
EMBO J., 19, 2000
|
|
1FD4
 
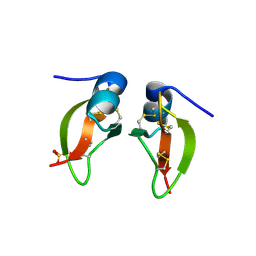 | | HUMAN BETA-DEFENSIN 2 | | Descriptor: | BETA-DEFENSIN 2, SULFATE ION | | Authors: | Hoover, D.M, Lubkowski, J. | | Deposit date: | 2000-07-19 | | Release date: | 2000-11-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of human beta-defensin-2 shows evidence of higher order oligomerization.
J.Biol.Chem., 275, 2000
|
|
1MAF
 
 | |
7GLQ
 
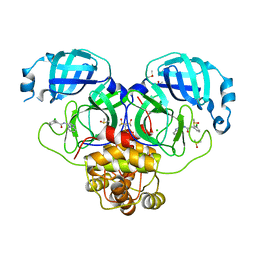 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with PET-UNK-8df914d1-2 (Mpro-P2007) | | Descriptor: | 2-(3-chlorophenyl)-N-[(4R)-imidazo[1,2-a]pyridin-3-yl]acetamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.063 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
1F95
 
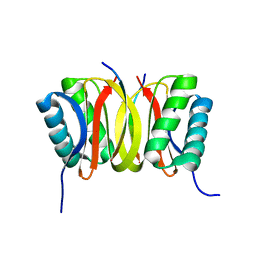 | | SOLUTION STRUCTURE OF DYNEIN LIGHT CHAIN 8 (DLC8) AND BIM PEPTIDE COMPLEX | | Descriptor: | BCL2-LIKE 11 (APOPTOSIS FACILITATOR), DYNEIN | | Authors: | Fan, J.-S, Zhang, Q, Tochio, H, Li, M, Zhang, M. | | Deposit date: | 2000-07-07 | | Release date: | 2001-02-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of diverse sequence-dependent target recognition by the 8 kDa dynein light chain.
J.Mol.Biol., 306, 2001
|
|
1FFQ
 
 | | CRYSTAL STRUCTURE OF CHITINASE A COMPLEXED WITH ALLOSAMIDIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITINASE A | | Authors: | Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K. | | Deposit date: | 2000-07-26 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo purification scheme and crystallization conditions yield high-resolution structures of chitinase A and its complex with the inhibitor allosamidin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1MLC
 
 | | MONOCLONAL ANTIBODY FAB D44.1 RAISED AGAINST CHICKEN EGG-WHITE LYSOZYME COMPLEXED WITH LYSOZYME | | Descriptor: | HEN EGG WHITE LYSOZYME, IGG1-KAPPA D44.1 FAB (HEAVY CHAIN), IGG1-KAPPA D44.1 FAB (LIGHT CHAIN) | | Authors: | Braden, B.C, Souchon, H, Eisele, J.-L, Bentley, G.A, Bhat, T.N, Navaza, J, Poljak, R.J. | | Deposit date: | 1995-03-10 | | Release date: | 1995-06-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structures of the free and the antigen-complexed Fab from monoclonal anti-lysozyme antibody D44.1.
J.Mol.Biol., 243, 1994
|
|
1MMK
 
 | | Crystal structure of ternary complex of the catalytic domain of human phenylalanine hydroxylase ((FeII)) complexed with tetrahydrobiopterin and thienylalanine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, BETA(2-THIENYL)ALANINE, FE (II) ION, ... | | Authors: | Andersen, O.A, Flatmark, T, Hough, E. | | Deposit date: | 2002-09-04 | | Release date: | 2003-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A resolution crystal structures of the ternary complexes of human phenylalanine hydroxylase catalytic domain with tetrahydrobiopterin and 3-(2-thienyl)-L-alanine or L-norleucine: substrate specificity and molecular motions related to substrate binding
J.Mol.Biol., 333, 2003
|
|
