5I8I
 
 | |
1J2P
 
 | | alpha-ring from the proteasome from archaeoglobus fulgidus | | Descriptor: | Proteasome alpha subunit | | Authors: | Groll, M, Brandstetter, H, Bartunik, H, Bourenkow, G, Huber, R. | | Deposit date: | 2003-01-08 | | Release date: | 2003-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigations on the Maturation and Regulation of Archaebacterial Proteasomes
J.MOL.BIOL., 327, 2003
|
|
1SSV
 
 | | Compensating bends in a 16 base-pair DNA oligomer containing a T3A3 segment | | Descriptor: | 5'-D(*CP*GP*AP*GP*GP*TP*TP*TP*AP*AP*AP*CP*CP*TP*CP*G)-3' | | Authors: | McAteer, K, Aceves-Gaona, A, Michalczyk, R, Buchko, G.W, Isern, N.G, Silks, L.A, Miller, J.H, Kennedy, M.A. | | Deposit date: | 2004-03-24 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Compensating bends in a 16-base-pair DNA oligomer containing a T(3)A(3) segment: A NMR study of global DNA curvature
Biopolymers, 75, 2004
|
|
1IRM
 
 | | Crystal structure of apo heme oxygenase-1 | | Descriptor: | apo heme oxygenase-1 | | Authors: | Sugishima, M, Sakamoto, H, Kakuta, Y, Omata, Y, Hayashi, S, Noguchi, M, Fukuyama, K. | | Deposit date: | 2001-10-09 | | Release date: | 2002-07-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of rat apo-heme oxygenase-1 (HO-1): mechanism of heme binding in HO-1 inferred from structural comparison of the apo and heme complex forms
Biochemistry, 41, 2002
|
|
1IS7
 
 | | Crystal structure of rat GTPCHI/GFRP stimulatory complex | | Descriptor: | GTP Cyclohydrolase I, GTP Cyclohydrolase I Feedback Regulatory Protein, PHENYLALANINE, ... | | Authors: | Maita, N, Okada, K, Hatakeyama, K, Hakoshima, T. | | Deposit date: | 2001-11-18 | | Release date: | 2002-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the stimulatory complex of GTP cyclohydrolase I and its feedback regulatory protein GFRP.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1T36
 
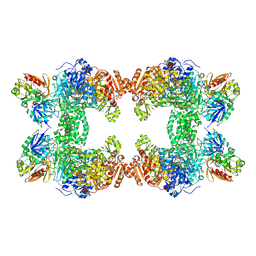 | | Crystal structure of E. coli carbamoyl phosphate synthetase small subunit mutant C248D complexed with uridine 5'-monophosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Carbamoyl-phosphate synthase large chain, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 2004-04-24 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Long-range allosteric transitions in carbamoyl phosphate synthetase.
Protein Sci., 13, 2004
|
|
1T1K
 
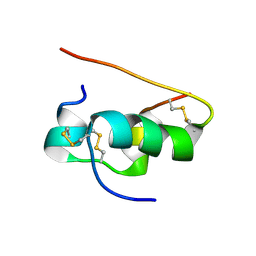 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT HIS-B10-ASP, VAL-B12-ALA, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | Insulin | | Authors: | Huang, K, Xu, B, Hu, S.Q, Chu, Y.C, Hua, Q.X, Whittaker, J, Nakagawa, S.H, De Meyts, P, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-10 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | How Insulin Binds: the B-Chain alpha-Helix Contacts the L1 beta-Helix of the Insulin Receptor.
J.Mol.Biol., 341, 2004
|
|
1IFC
 
 | |
1IGZ
 
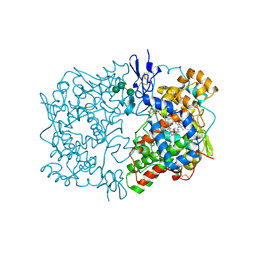 | | Crystal Structure of Linoleic acid Bound in the Cyclooxygenase Channel of Prostaglandin Endoperoxide H Synthase-1. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Malkowski, M.G, Thuresson, E.D, Smith, W.L, Garavito, R.M. | | Deposit date: | 2001-04-18 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of eicosapentaenoic and linoleic acids in the cyclooxygenase site of prostaglandin endoperoxide H synthase-1.
J.Biol.Chem., 276, 2001
|
|
5ILW
 
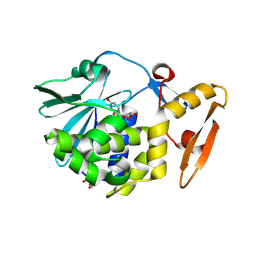 | | Crystal structure of the complex of type 1 Ribosome inactivating protein from Momordica balsamina with Uridine at 1.97 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Singh, P.K, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-03-05 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein from Momordica balsamina with uracil and uridine.
Proteins, 87, 2019
|
|
5IM4
 
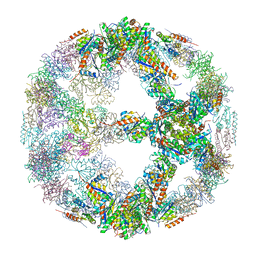 | | Crystal structure of designed two-component self-assembling icosahedral cage I52-32 | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, Phosphotransferase system, mannose/fructose-specific component IIA | | Authors: | Liu, Y.A, Cascio, D, Sawaya, M.R, Bale, J.B, Collazo, M.J, Thomas, C, Sheffler, W, King, N.P, Baker, D, Yeates, T.O. | | Deposit date: | 2016-03-05 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Accurate design of megadalton-scale two-component icosahedral protein complexes.
Science, 353, 2016
|
|
1TDW
 
 | | Crystal structure of double truncated human phenylalanine hydroxylase BH4-responsive PKU mutant A313T. | | Descriptor: | FE (III) ION, Phenylalanine-4-hydroxylase | | Authors: | Erlandsen, H, Pey, A.L, Gamez, A, Perez, B, Desviat, L.R, Aguado, C, Koch, R, Surendran, S, Tyring, S, Matalon, R, Scriver, C.R, Ugarte, M, Martinez, A, Stevens, R.C. | | Deposit date: | 2004-05-24 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Correction of kinetic and stability defects by tetrahydrobiopterin in phenylketonuria patients with certain phenylalanine hydroxylase mutations.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
5IPI
 
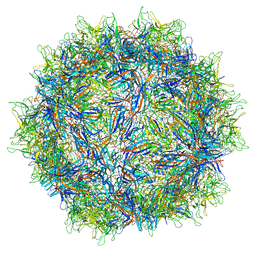 | | Structure of Adeno-associated virus type 2 VLP | | Descriptor: | Capsid protein VP1 | | Authors: | Drouin, L.M, Lins, B, Janssen, M.E, Bennet, A, Chipman, P, McKenna, R, Chen, W, Muzyczka, N, Cardone, G, Baker, T.S, Agbandje-McKenna, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-electron Microscopy Reconstruction and Stability Studies of the Wild Type and the R432A Variant of Adeno-associated Virus Type 2 Reveal that Capsid Structural Stability Is a Major Factor in Genome Packaging.
J.Virol., 90, 2016
|
|
1J2E
 
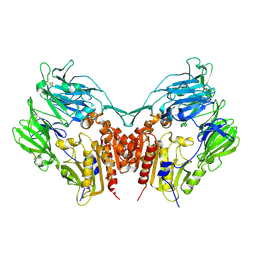 | | Crystal structure of Human Dipeptidyl peptidase IV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV | | Authors: | Hiramatsu, H, Kyono, K, Higashiyama, Y, Fukushima, C, Shima, H, Sugiyama, S, Inaka, K, Yamamoto, A, Shimizu, R. | | Deposit date: | 2002-12-30 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure and function of human dipeptidyl peptidase IV, possessing a unique eight-bladed beta-propeller fold.
Biochem.Biophys.Res.Commun., 302, 2003
|
|
5I14
 
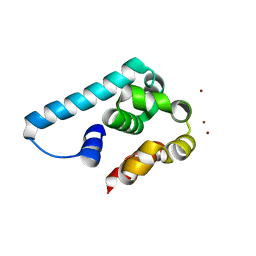 | | Truncated and mutated T4 lysozyme | | Descriptor: | NICKEL (II) ION, mutated and truncated T4 lysozyme | | Authors: | Klima, M, Boura, E. | | Deposit date: | 2016-02-05 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.745 Å) | | Cite: | Metal ions-binding T4 lysozyme as an intramolecular protein purification tag compatible with X-ray crystallography.
Protein Sci., 26, 2017
|
|
5I1D
 
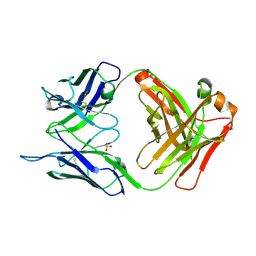 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV3-23/IGKV4-1 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5IRM
 
 | |
5IS4
 
 | |
1IMD
 
 | |
1T16
 
 | | Crystal structure of the bacterial fatty acid transporter FadL from Escherichia coli | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, COPPER (II) ION, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | van den Berg, B, Black, P.N, Clemons Jr, W.M, Rapoport, T.A. | | Deposit date: | 2004-04-15 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the long-chain fatty acid transporter FadL.
Science, 304, 2004
|
|
5IJZ
 
 | |
5I6R
 
 | | Crystal Structure of srGAP2 F-BARx WT Form-1 | | Descriptor: | ACETATE ION, D-MALATE, SLIT-ROBO Rho GTPase-activating protein 2, ... | | Authors: | Sporny, M, Guez-Haddad, J, Isupov, M.N, Opatowsky, Y. | | Deposit date: | 2016-02-16 | | Release date: | 2017-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for srGAP2 Membrane Interactions, and Antagonism by the Human Specific Paralog srGAP2C
To Be Published
|
|
1IML
 
 | | CYSTEINE RICH INTESTINAL PROTEIN, NMR, 48 STRUCTURES | | Descriptor: | CYSTEINE RICH INTESTINAL PROTEIN, ZINC ION | | Authors: | Perez-Alvarado, G.C, Kosa, J.L, Louis, H.A, Beckerle, M.C, Winge, D.R, Summers, M.F. | | Deposit date: | 1995-12-23 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the cysteine-rich intestinal protein, CRIP.
J.Mol.Biol., 257, 1996
|
|
5IL7
 
 | |
1T3Q
 
 | | Crystal structure of quinoline 2-Oxidoreductase from Pseudomonas Putida 86 | | Descriptor: | DIOXOSULFIDOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bonin, I, Martins, B.M, Purvanov, V, Fetzner, S, Huber, R, Dobbek, H. | | Deposit date: | 2004-04-27 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Active site geometry and substrate recognition of the molybdenum hydroxylase quinoline 2-oxidoreductase.
STRUCTURE, 12, 2004
|
|
