1EOT
 
 | |
1HB5
 
 | | quasi-atomic resolution model of bacteriophage PRD1 P3-shell, obtained by combined cryo-EM and X-ray crystallography. | | Descriptor: | BACTERIOPHAGE PRD1 P3-SHELL | | Authors: | San Martin, C, Burnett, R.M, De Haas, F, Heinkel, R, Rutten, T, Fuller, S.D, Butcher, S.J, Bamford, D.H. | | Deposit date: | 2001-04-11 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Combined Em/X-Ray Imaging Yields a Quasi-Atomic Model of the Adenovirus-Related Bacteriophage Prd1 and Shows Key Capsid and Membrane Interactions.
Structure, 9, 2001
|
|
7X14
 
 | | Crystal structure of phospho-FFAT motif of MIGA2 bound to VAPB | | Descriptor: | MIGA2 phospho FFAT motif, SULFATE ION, Vesicle-associated membrane protein-associated protein B | | Authors: | Kim, H, Lee, C. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.675 Å) | | Cite: | Structural basis for mitoguardin-2 mediated lipid transport at ER-mitochondrial membrane contact sites.
Nat Commun, 13, 2022
|
|
1FU2
 
 | | FIRST PROTEIN STRUCTURE DETERMINED FROM X-RAY POWDER DIFFRACTION DATA | | Descriptor: | CHLORIDE ION, INSULIN, A CHAIN, ... | | Authors: | Von Dreele, R.B, Stephens, P.W, Blessing, R.H, Smith, G.D. | | Deposit date: | 2000-09-13 | | Release date: | 2000-10-16 | | Last modified: | 2018-10-03 | | Method: | POWDER DIFFRACTION | | Cite: | The first protein crystal structure determined from high-resolution X-ray powder diffraction data: a variant of T3R3 human insulin-zinc complex produced by grinding.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
6K51
 
 | |
1TMJ
 
 | | Crystal structure of E.coli apo-HPPK(W89A) at 1.45 Angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Blaszczyk, J, Ji, X. | | Deposit date: | 2004-06-10 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Is the Critical Role of Loop 3 of Escherichia coli 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase in Catalysis Due to Loop-3 Residues Arginine-84 and Tryptophan-89? Site-Directed Mutagenesis, Biochemical, and Crystallographic Studies.
Biochemistry, 44, 2005
|
|
6KM8
 
 | | Crystal Structure of Momordica charantia 7S globulin | | Descriptor: | 7S globulin, ACETATE ION, COPPER (II) ION | | Authors: | Kesari, P, Pratap, S, Dhankhar, P, Dalal, V, Kumar, P. | | Deposit date: | 2019-07-31 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Structural characterization and in-silico analysis of Momordica charantia 7S globulin for stability and ACE inhibition.
Sci Rep, 10, 2020
|
|
2I6T
 
 | | Orthorhombic Structure of the LDH domain of Human Ubiquitin-conjugating Enzyme E2-like Isoform A | | Descriptor: | GLYCEROL, SULFATE ION, UBIQUITIN-CONJUGATING ENZYME E2-LIKE ISOFORM A | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Finerty Jr, P.J, Butler-Cole, C, Tempel, W, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S. | | Deposit date: | 2006-08-29 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Investigation into the L-lactate Dehydrogenase Domain of Human Ubiquitin-conjugating Enzyme E2-like Isoform A
To be Published
|
|
6K50
 
 | |
3AZE
 
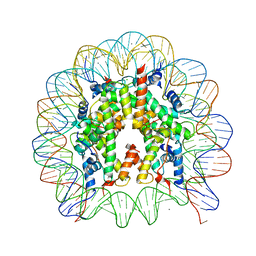 | | Crystal Structure of Human Nucleosome Core Particle Containing H3K64Q mutation | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-05-25 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
3AZG
 
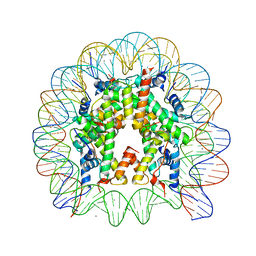 | | Crystal Structure of Human Nucleosome Core Particle Containing H3K115Q mutation | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-05-25 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
2XHL
 
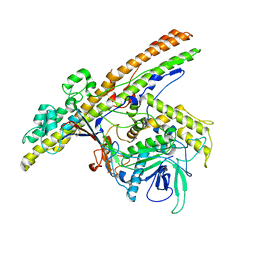 | | Structure of a functional derivative of Clostridium botulinum neurotoxin type B | | Descriptor: | BOTULINUM NEUROTOXIN B HEAVY CHAIN, BOTULINUM NEUROTOXIN B LIGHT CHAIN, ZINC ION | | Authors: | Masuyer, G, Beard, M, Cadd, V.A, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2010-06-18 | | Release date: | 2010-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and Activity of a Functional Derivative of Clostridium Botulinum Neurotoxin B.
J.Struct.Biol., 174, 2011
|
|
3AZJ
 
 | | Crystal Structure of Human Nucleosome Core Particle Containing H4K44Q mutation | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-05-25 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
3AZK
 
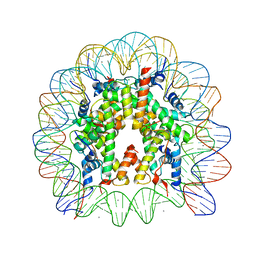 | | Crystal Structure of Human Nucleosome Core Particle Containing H4K59Q mutation | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-05-25 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
4M0U
 
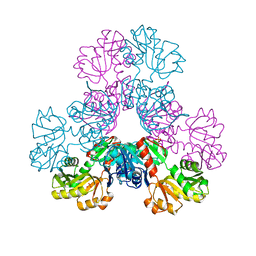 | | crystal structure of human PRS1 Q133P mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-08-02 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
1EQ8
 
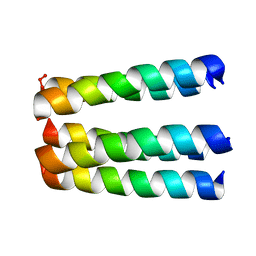 | | THREE-DIMENSIONAL STRUCTURE OF THE PENTAMERIC HELICAL BUNDLE OF THE ACETYLCHOLINE RECEPTOR M2 TRANSMEMBRANE SEGMENT | | Descriptor: | ACETYLCHOLINE RECEPTOR PROTEIN | | Authors: | Marassi, F.M, Gesell, J.J, Kim, Y, Valente, A.P, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 2000-04-03 | | Release date: | 2000-04-26 | | Last modified: | 2024-09-25 | | Method: | SOLID-STATE NMR | | Cite: | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
2JIR
 
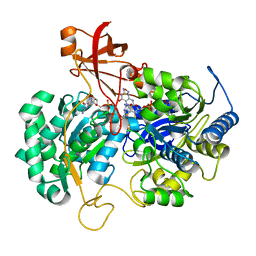 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CYANIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J.G, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
2F36
 
 | |
1F0Z
 
 | | SOLUTION STRUCTURE OF THIS, THE SULFUR CARRIER PROTEIN IN E.COLI THIAMIN BIOSYNTHESIS | | Descriptor: | THIS PROTEIN | | Authors: | Wang, C, Xi, J, Begley, T.P, Nicholson, L.K. | | Deposit date: | 2000-05-17 | | Release date: | 2001-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ThiS and implications for the evolutionary roots of ubiquitin.
Nat.Struct.Biol., 8, 2001
|
|
2H3O
 
 | | Structure of MERFT, a membrane protein with two trans-membrane helices | | Descriptor: | MerF | | Authors: | Opella, S.J, De Angelis, A.A, Howell, S.C, Nevzorov, A.A. | | Deposit date: | 2006-05-22 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-29 | | Method: | SOLID-STATE NMR | | Cite: | Structure Determination of a Membrane Protein with Two Trans-membrane Helices in Aligned Phospholipid Bicelles by Solid-State NMR Spectroscopy.
J.Am.Chem.Soc., 128, 2006
|
|
2DTM
 
 | | Thermodynamic and structural analyses of hydrolytic mechanism by catalytic antibodies | | Descriptor: | IMMUNOGLOBULIN 6D9 | | Authors: | Oda, M, Ito, N, Tsumuraya, T, Suzuki, K, Fujii, I. | | Deposit date: | 2006-07-13 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
3C13
 
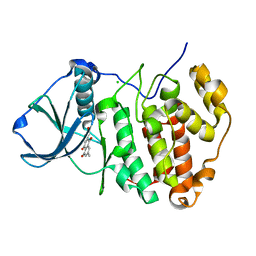 | | Low pH-value crystal structure of emodin in complex with the catalytic subunit of protein kinase CK2 | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Niefind, K, Raaf, J, Issinger, O.-G. | | Deposit date: | 2008-01-22 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Catalytic Subunit of Human Protein Kinase CK2 Structurally Deviates from Its Maize Homologue in Complex with the Nucleotide Competitive Inhibitor Emodin
J.Mol.Biol., 377, 2008
|
|
3C9A
 
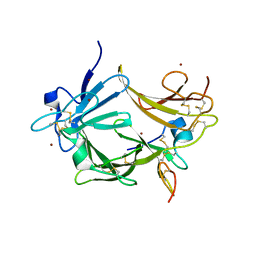 | | High Resolution Crystal Structure of Argos bound to the EGF domain of Spitz | | Descriptor: | BROMIDE ION, Protein giant-lens, Protein spitz | | Authors: | Klein, D.E, Stayrook, S.E, Shi, F, Narayan, K, Lemmon, M.A. | | Deposit date: | 2008-02-15 | | Release date: | 2008-05-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for EGFR ligand sequestration by Argos.
Nature, 453, 2008
|
|
2FBN
 
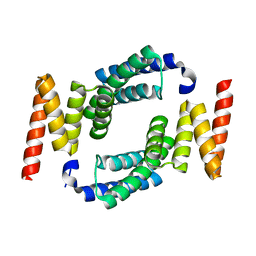 | | Plasmodium falciparum putative FK506-binding protein PFL2275c, C-terminal TPR-containing domain | | Descriptor: | 70 kDa peptidylprolyl isomerase, putative | | Authors: | Dong, A, Lew, J, Koeieradzki, I, Sundararajan, E, Melone, M, Wasney, G, Zhao, Y, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-12-09 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystallographic structure of the tetratricopeptide repeat domain of Plasmodium falciparum FKBP35 and its molecular interaction with Hsp90 C-terminal pentapeptide.
Protein Sci., 18, 2009
|
|
2F34
 
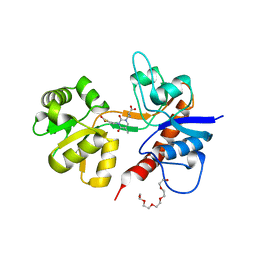 | |
