8GCE
 
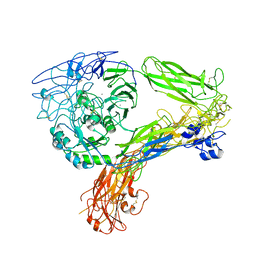 | | The Extracellular Domain of Integrin AlphaIIbBeta3 in Intermediate State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huo, T, Wu, H, Wang, Z. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Full-length alpha IIb beta 3 cryo-EM structure reveals intact integrin initiate-activation intrinsic architecture.
Structure, 32, 2024
|
|
8GCD
 
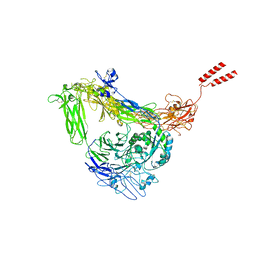 | | Full length Integrin AlphaIIbBeta3 in inactive state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huo, T, Wu, H, Wang, Z. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Full-length alpha IIb beta 3 cryo-EM structure reveals intact integrin initiate-activation intrinsic architecture.
Structure, 32, 2024
|
|
8CEH
 
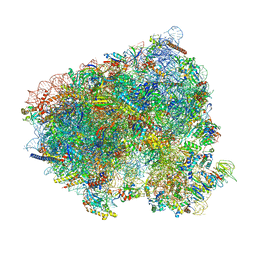 | | Translocation intermediate 4 (TI-4) of 80S S. cerevisiae ribosome with ligands and eEF2 in the presence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
5HI4
 
 | | Binding site elucidation and structure guided design of macrocyclic IL-17A antagonists | | Descriptor: | (9'S,17'R)-6'-chloro-N-methyl-9'-{[(1-methyl-1H-pyrazol-5-yl)carbonyl]amino}-10',19'-dioxo-2'-oxa-11',18'-diazaspiro[cyclopentane-1,21'-tetracyclo[20.2.2.2~12,15~.1~3,7~]nonacosane]-1'(24'),3'(29'),4',6',12',14',22',25',27'-nonaene-17'-carboxamide, CAT-2000 FAB heavy chain, CAT-2000 FAB light chain, ... | | Authors: | Liu, S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site elucidation and structure guided design of macrocyclic IL-17A antagonists.
Sci Rep, 6, 2016
|
|
1RX6
 
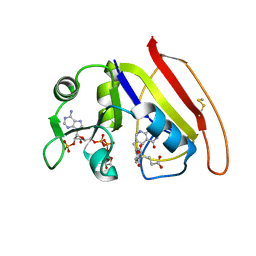 | |
6QKZ
 
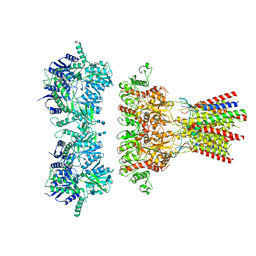 | | Full length GluA1/2-gamma8 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-sulfonamide, ... | | Authors: | Herguedas, B, Garcia-Nafria, J, Greger, I.G. | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP gamma 8.
Science, 364, 2019
|
|
6FFE
 
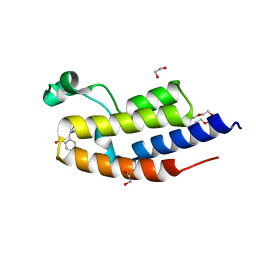 | |
3WSY
 
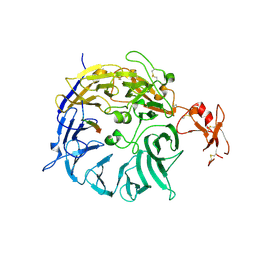 | | SorLA Vps10p domain in complex with its own propeptide fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin-related receptor, peptide from Sortilin-related receptor | | Authors: | Kitago, Y, Nakata, Z, Nagae, M, Nogi, T, Takagi, J. | | Deposit date: | 2014-03-30 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for amyloidogenic peptide recognition by sorLA.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6R75
 
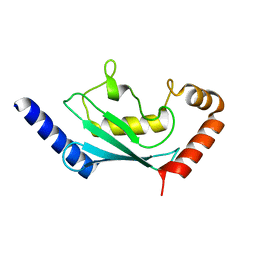 | | Crystal structure of human Ube2T E54R mutant | | Descriptor: | Ubiquitin-conjugating enzyme E2 T | | Authors: | Chaugule, V.K, Rennie, M.L, Walden, H, Arkinson, C, Kamarainen, O, Toth, R. | | Deposit date: | 2019-03-28 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric mechanism for site-specific ubiquitination of FANCD2.
Nat.Chem.Biol., 16, 2020
|
|
4FZJ
 
 | | Pantothenate synthetase in complex with 1,3-DIMETHYL-1H-THIENO[2,3-C]PYRAZOLE-5-CARBOXYLIC ACID | | Descriptor: | 1,2-ETHANEDIOL, 1,3-dimethyl-1H-thieno[2,3-c]pyrazole-5-carboxylic acid, ETHANOL, ... | | Authors: | Silvestre, H.L, Blundell, T.L, Abell, C, Ciulli, A. | | Deposit date: | 2012-07-06 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Integrated biophysical approach to fragment screening and validation for fragment-based lead discovery.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1ZNS
 
 | | Crystal structure of N-ColE7/12-bp DNA/Zn complex | | Descriptor: | 5'-D(*CP*GP*GP*GP*AP*TP*AP*TP*CP*CP*CP*G)-3', Colicin E7, ZINC ION | | Authors: | Doudeva, L.G, Huang, H, Hsia, K.C, Shi, Z, Li, C.L, Shen, Y, Yuan, H.S. | | Deposit date: | 2005-05-12 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structural analysis and metal-dependent stability and activity studies of the ColE7 endonuclease domain in complex with DNA/Zn2+ or inhibitor/Ni2+
Protein Sci., 15, 2006
|
|
8CDR
 
 | | Translocation intermediate 2 (TI-2) of 80S S. cerevisiae ribosome with ligands and eEF2 in the presence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-01-31 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.04 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
8CF5
 
 | | Translocation intermediate 1 (TI-1) of 80S S. cerevisiae ribosome with ligands and eEF2 in the presence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-02 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
8CCS
 
 | | 80S S. cerevisiae ribosome with ligands in hybrid-1 pre-translocation (PRE-H1) complex | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-01-27 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (1.97 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
8CKU
 
 | | Translocation intermediate 1 (TI-1*) of 80S S. cerevisiae ribosome with ligands and eEF2 in the absence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-16 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
8CIV
 
 | | Translocation intermediate 5 (TI-5) of 80S S. cerevisiae ribosome with ligands and eEF2 in the presence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
1RF7
 
 | | STRUCTURE OF DIHYDROFOLATE REDUCTASE COMPLEXED WITH DIHYDROFOLATE | | Descriptor: | CHLORIDE ION, DIHYDROFOLATE REDUCTASE, DIHYDROFOLIC ACID, ... | | Authors: | Sawaya, M.R. | | Deposit date: | 1996-10-24 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence.
Biochemistry, 36, 1997
|
|
1RG7
 
 | | DIHYDROFOLATE REDUCTASE COMPLEXED WITH METHOTREXATE | | Descriptor: | DIHYDROFOLATE REDUCTASE, METHOTREXATE | | Authors: | Sawaya, M.R, Kraut, J. | | Deposit date: | 1996-11-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence.
Biochemistry, 36, 1997
|
|
1RH3
 
 | |
1G9F
 
 | | CRYSTAL STRUCTURE OF THE SOYBEAN AGGLUTININ IN A COMPLEX WITH A BIANTENNARY BLOOD GROUP ANTIGEN ANALOG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LECTIN, ... | | Authors: | Buts, L, Hamelryck, T.W, Dao-Thi, M.-H, Loris, R, Wyns, L, Etzler, M.E. | | Deposit date: | 2000-11-23 | | Release date: | 2001-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Weak protein-protein interactions in lectins: the crystal structure of a vegetative lectin from the legume Dolichos biflorus.
J.Mol.Biol., 309, 2001
|
|
8H21
 
 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with L-alanine | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, Serine palmitoyltransferase | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8H1Y
 
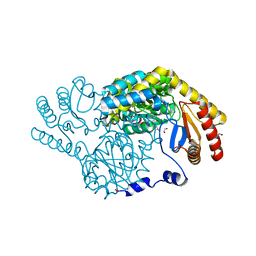 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with L-homoserine | | Descriptor: | (2~{S})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-4-oxidanyl-butanoic acid, 1,2-ETHANEDIOL, Serine palmitoyltransferase | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
7EQT
 
 | |
7EQS
 
 | |
7ER1
 
 | |
