6XZO
 
 | | Crystal structure of human carbonic anhydrase I in complex with 4-(3-(2-((4-bromo-2-hydroxybenzyl)amino)ethyl)ureido) benzenesulfonamide | | Descriptor: | 1-[2-[(4-bromanyl-2-oxidanyl-phenyl)methylamino]ethyl]-3-(4-sulfamoylphenyl)urea, Carbonic anhydrase 1, ZINC ION | | Authors: | Zanotti, G, Majid, A, Bozdag, M, Angeli, A, Carta, F, Berto, P, Supuran, C. | | Deposit date: | 2020-02-05 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Benzylaminoethyureido-Tailed Benzenesulfonamides: Design, Synthesis, Kinetic and X-ray Investigations on Human Carbonic Anhydrases.
Int J Mol Sci, 21, 2020
|
|
4U0X
 
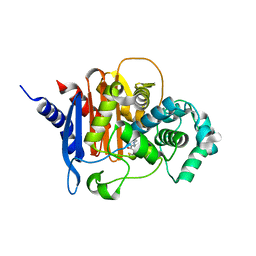 | | Structure of ADC-7 beta-lactamase in complex with boronic acid inhibitor S02030 | | Descriptor: | 1-{(2R)-2-(dihydroxyboranyl)-2-[(thiophen-2-ylacetyl)amino]ethyl}-1H-1,2,3-triazole-4-carboxylic acid, ADC-7 beta-lactamase, PHOSPHATE ION | | Authors: | Powers, R.A, Wallar, B.J, Swanson, H.C. | | Deposit date: | 2014-07-14 | | Release date: | 2014-11-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Biochemical and Structural Analysis of Inhibitors Targeting the ADC-7 Cephalosporinase of Acinetobacter baumannii.
Biochemistry, 53, 2014
|
|
1FG5
 
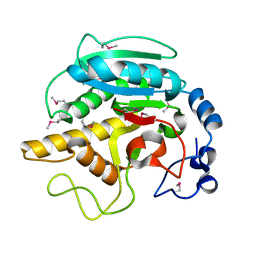 | | CRYSTAL STRUCTURE OF BOVINE ALPHA-1,3-GALACTOSYLTRANSFERASE CATALYTIC DOMAIN. | | Descriptor: | N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE | | Authors: | Gastinel, L.N, Bignon, C, Shaper, J.H, Joziasse, D.H. | | Deposit date: | 2000-07-28 | | Release date: | 2001-07-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bovine alpha1,3-galactosyltransferase catalytic domain structure and its relationship with ABO histo-blood group and glycosphingolipid glycosyltransferases.
EMBO J., 20, 2001
|
|
4Y22
 
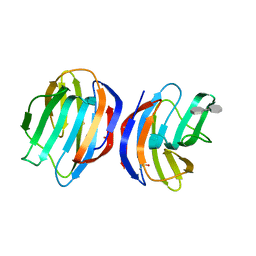 | | Complex of human Galectin-1 and (3OSO3)Galbeta1-3GlcNAc | | Descriptor: | 3-O-sulfo-beta-D-galactopyranose-(1-3)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Galectin-1 | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
8GFV
 
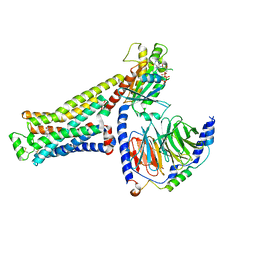 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #1 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8USU
 
 | | Crystal Structure of L-galactose 1-dehydrogenase of Myrciaria dubia in complex with NAD | | Descriptor: | L-galactose dehydrogenase isoform X1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2023-10-30 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit camu-camu.
J.Exp.Bot., 75, 2024
|
|
4TXR
 
 | |
8UOW
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21A) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Asparaginase | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
1FGI
 
 | |
6XRU
 
 | | GTP-specific succinyl-CoA synthetase complexed with desulfo-coenzyme A, magnesium ions and succinates | | Descriptor: | 1,2-ETHANEDIOL, DESULFO-COENZYME A, GLYCEROL, ... | | Authors: | Huang, J, Fraser, M.E. | | Deposit date: | 2020-07-13 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Second distinct conformation of the phosphohistidine loop in succinyl-CoA synthetase
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7A1B
 
 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the ATP-competitive inhibitor 5,6-dibromo-1H-triazolo[4,5-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, 5,6-dibromo-1H-triazolo[4,5-b]pyridine, CHLORIDE ION, ... | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-12 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.287 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
8AYJ
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiens complexed with 3-aminooxypropionic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
4UCR
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-hydroxy-2-methylquinoline-6-carboxamide, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
8PO2
 
 | |
7AB2
 
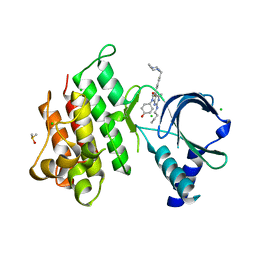 | | Crystal structure of MerTK kinase domain in complex with UNC2025 | | Descriptor: | 4-[2-(butylamino)-5-[4-[(4-methylpiperazin-1-yl)methyl]phenyl]pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexan-1-ol, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
7A6B
 
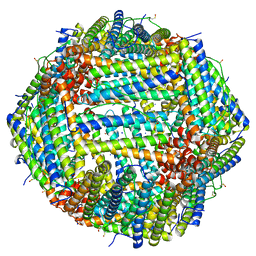 | | 1.33 A structure of human apoferritin obtained from Titan Mono- BCOR microscope | | Descriptor: | Ferritin heavy chain, SODIUM ION | | Authors: | Yip, K.M, Fischer, N, Chari, A, Stark, H. | | Deposit date: | 2020-08-25 | | Release date: | 2020-09-02 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (1.33 Å) | | Cite: | Atomic-resolution protein structure determination by cryo-EM.
Nature, 587, 2020
|
|
6X95
 
 | |
7AB1
 
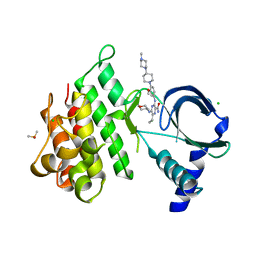 | | Crystal structure of MerTK kinase domain in complex with Gilteritinib | | Descriptor: | 6-ethyl-3-[[3-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]amino]-5-(oxan-4-ylamino)pyrazine-2-carboxamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
1FH7
 
 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED INHIBITOR DEOXYNOJIRIMYCIN | | Descriptor: | BETA-1,4-XYLANASE, PIPERIDINE-3,4,5-TRIOL, beta-D-xylopyranose | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
6X9M
 
 | |
8VLJ
 
 | | Crystal structure of the cacodylate-bound yeast cytosine deaminase (closed form) | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, Cytosine deaminase, ... | | Authors: | Picard, M.-E, Grenier, J, Despres, P.C, Dube, A.K, Landry, C.R, Shi, R. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Compensatory mutations potentiate constructive neutral evolution by gene duplication.
Science, 385, 2024
|
|
6ZYQ
 
 | | Structure of NDM-1 with 2-Mercaptomethyl-thiazolidine D-syn-1b | | Descriptor: | (2~{S},4~{S})-2-ethoxycarbonyl-5,5-dimethyl-2-(sulfanylmethyl)-1,3-thiazolidine-4-carboxylic acid, Metallo-beta-lactamase type 2, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 2-Mercaptomethyl-thiazolidines use conserved aromatic-S interactions to achieve broad-range inhibition of metallo-beta-lactamases.
Chem Sci, 12, 2021
|
|
1FHX
 
 | | Structure of the pleckstrin homology domain from GRP1 in complex with inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | GUANINE NUCLEOTIDE EXCHANGE FACTOR AND INTEGRIN BINDING PROTEIN HOMOLOG GRP1, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, SULFATE ION | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-08-02 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains
Mol.Cell, 6, 2000
|
|
6X7J
 
 | |
6X7Y
 
 | |
