7W9V
 
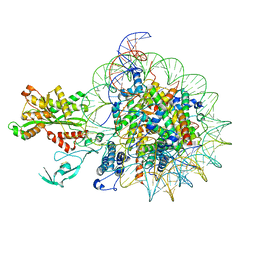 | | Cryo-EM structure of nucleosome in complex with p300 acetyltransferase catalytic core (complex I) | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Hatazawa, S, Liu, J, Takizawa, Y, Zandian, M, Negishi, L, Kutateladze, T.G, Kurumizaka, H. | | Deposit date: | 2021-12-10 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural basis for binding diversity of acetyltransferase p300 to the nucleosome.
Iscience, 25, 2022
|
|
6N8T
 
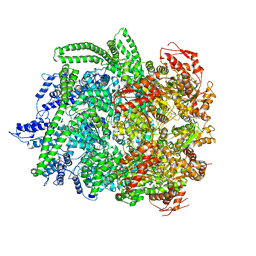 | | Hsp104DWB closed conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
8KGW
 
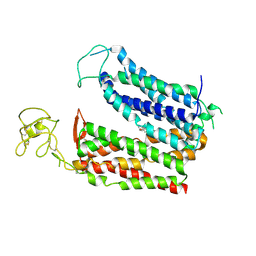 | | Molecular mechanism of prostaglandin transporter SLCO2A1 | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Solute carrier organic anion transporter family member 2A1 | | Authors: | Li, Y.H, Qu, Q.H, Zhu, Z.N, Chao, Y.L, Zhou, Z.X. | | Deposit date: | 2023-08-19 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular mechanism of prostaglandin transporter SLCO2A1
To Be Published
|
|
6ND4
 
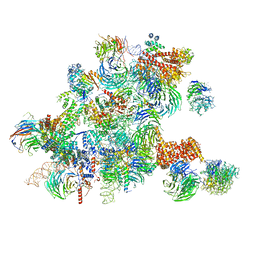 | | Conformational switches control early maturation of the eukaryotic small ribosomal subunit | | Descriptor: | 18S rRNA 5' domain start, 5'ETS rRNA, Bud21, ... | | Authors: | Hunziker, M, Barandun, J, Klinge, S. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Conformational switches control early maturation of the eukaryotic small ribosomal subunit.
Elife, 8, 2019
|
|
5IV3
 
 | | Crystal structure of human soluble adenylyl cyclase in complex with alpha,beta-methyleneadenosine-5'-triphosphate and the allosteric inhibitor LRE1 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-N~4~-cyclopropyl-N~4~-[(thiophen-2-yl)methyl]pyrimidine-2,4-diamine, ACETATE ION, ... | | Authors: | Kleinboelting, S, Steegborn, C. | | Deposit date: | 2016-03-18 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of LRE1 as a specific and allosteric inhibitor of soluble adenylyl cyclase.
Nat.Chem.Biol., 12, 2016
|
|
6NBB
 
 | |
1Y20
 
 | |
5MKF
 
 | | cryoEM Structure of Polycystin-2 in complex with calcium and lipids | | Descriptor: | 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wilkes, M, Madej, M.G, Ziegler, C. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular insights into lipid-assisted Ca(2+) regulation of the TRP channel Polycystin-2.
Nat. Struct. Mol. Biol., 24, 2017
|
|
8OX4
 
 | | Cryo-EM structure of ATP8B1-CDC50A in E1-ATP conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell cycle control protein 50A, MAGNESIUM ION, ... | | Authors: | Dieudonne, T, Kummerer, F, Juknaviciute Laursen, M, Stock, C, Kock Flygaard, R, Khalid, S, Lenoir, G, Lyons, J.A, Lindorff-Larsen, K, Nissen, P. | | Deposit date: | 2023-05-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Activation and substrate specificity of the human P4-ATPase ATP8B1.
Nat Commun, 14, 2023
|
|
1Y1M
 
 | |
1Y1Z
 
 | |
1JLN
 
 | | Crystal structure of the catalytic domain of protein tyrosine phosphatase PTP-SL/BR7 | | Descriptor: | Protein Tyrosine Phosphatase, receptor type, R | | Authors: | Szedlacsek, S.E, Aricescu, A.R, Fulga, T.A, Renault, L, Scheidig, A.J. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of PTP-SL/PTPBR7 catalytic domain: implications for MAP kinase regulation.
J.Mol.Biol., 311, 2001
|
|
8P94
 
 | |
8HWU
 
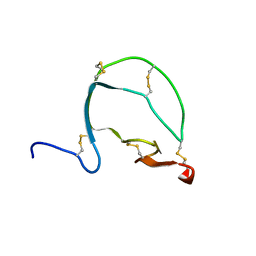 | | Solution structure of frog peptide LL-TIL | | Descriptor: | LL-TIL | | Authors: | Rami, M.J, Sarma, S.P. | | Deposit date: | 2023-01-03 | | Release date: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural, Functional, and Mutational Studies of a Potent Subtilisin Inhibitor from Budgett's Frog, Lepidobatrachus laevis.
Biochemistry, 62, 2023
|
|
6NR3
 
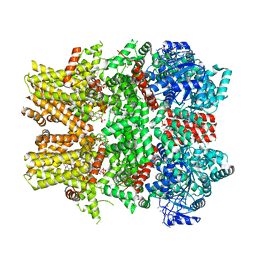 | | Cryo-EM structure of the TRPM8 ion channel in complex with high occupancy icilin, PI(4,5)P2, and calcium | | Descriptor: | (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl icosa-5,8,11,14-tetraenoate, CALCIUM ION, Icilin, ... | | Authors: | Yin, Y, Le, S.C, Hsu, A.L, Borgnia, M.J, Yang, H, Lee, S.-Y. | | Deposit date: | 2019-01-22 | | Release date: | 2019-02-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel.
Science, 363, 2019
|
|
6TAX
 
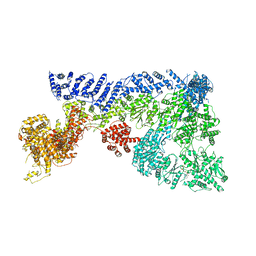 | | Mouse RNF213 wild type protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNF213,E3 ubiquitin-protein ligase RNF213,E3 ubiquitin-protein ligase RNF213, ... | | Authors: | Ahel, J, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-01 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Moyamoya disease factor RNF213 is a giant E3 ligase with a dynein-like core and a distinct ubiquitin-transfer mechanism.
Elife, 9, 2020
|
|
8PB9
 
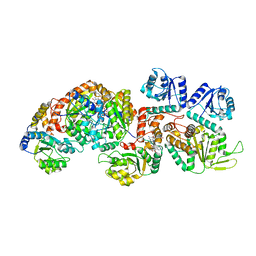 | | Cryo-EM structure of the c-di-GMP-bound FleQ-FleN master regulator complex from Pseudomonas aeruginosa | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Antiactivator FleN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Torres-Sanchez, L.T, Krasteva, P.V. | | Deposit date: | 2023-06-08 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the P. aeruginosa FleQ-FleN master regulators reveal large-scale conformational switching in motility and biofilm control.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PKD
 
 | |
6BV7
 
 | |
6MZM
 
 | | Human TFIID bound to promoter DNA and TFIIA | | Descriptor: | SCP DNA (80-MER), TATA-box-binding protein, Transcription initiation factor IIA subunit 1, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
6SKO
 
 | | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork - conformation 2 MCM CTD:ssDNA | | Descriptor: | DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, DNA replication licensing factor MCM4, ... | | Authors: | Yeeles, J, Baretic, D, Jenkyn-Bedford, M. | | Deposit date: | 2019-08-16 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork.
Mol.Cell, 78, 2020
|
|
6Z41
 
 | |
6N0F
 
 | |
6TED
 
 | | Structure of complete, activated transcription complex Pol II-DSIF-PAF-SPT6 uncovers allosteric elongation activation by RTF1 | | Descriptor: | DNA (37-MER), DNA-directed RNA polymerase II subunit RPB9, DNA-directed RNA polymerase subunit, ... | | Authors: | Vos, S.M, Farnung, L, Cramer, P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of complete Pol II-DSIF-PAF-SPT6 transcription complex reveals RTF1 allosteric activation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6NXE
 
 | |
