5IXH
 
 | | Crystal Structure of Burkholderia cenocepacia BcnA | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, ... | | Authors: | Loutet, S.A, Murphy, M.E.P. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Antibiotic Capture by Bacterial Lipocalins Uncovers an Extracellular Mechanism of Intrinsic Antibiotic Resistance.
MBio, 8, 2017
|
|
7K58
 
 | |
7K5B
 
 | |
7KEK
 
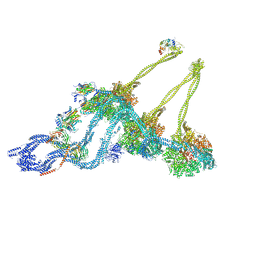 | | Structure of the free outer-arm dynein in pre-parallel state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Dynein alpha heavy chain, ... | | Authors: | Rao, Q, Zhang, K. | | Deposit date: | 2020-10-11 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structures of outer-arm dynein array on microtubule doublet reveal a motor coordination mechanism.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3FQA
 
 | | Gabaculien complex of gabaculine resistant GSAM version | | Descriptor: | 3-AMINOBENZOIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Glutamate-1-semialdehyde 2,1-aminomutase | | Authors: | Stetefeld, J. | | Deposit date: | 2009-01-07 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Absence of a catalytic water confers resistance to the neurotoxin gabaculine.
Faseb J., 24, 2010
|
|
3FQ7
 
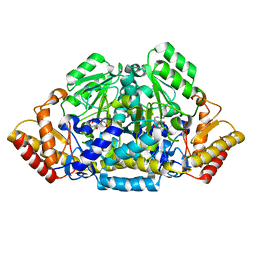 | | Gabaculine complex of GSAM | | Descriptor: | 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, Glutamate-1-semialdehyde 2,1-aminomutase | | Authors: | Stetefeld, J. | | Deposit date: | 2009-01-07 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Absence of a catalytic water confers resistance to the neurotoxin gabaculine.
Faseb J., 24, 2010
|
|
6FRR
 
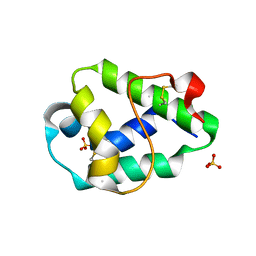 | | Structural and immunological properties of the allergen Art v 3 | | Descriptor: | Non-specific lipid-transfer protein, SULFATE ION | | Authors: | Brandstetter, H, Soh, W.T, Magler, I. | | Deposit date: | 2018-02-16 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Boiling down the cysteine-stabilized LTP fold - loss of structural and immunological integrity of allergenic Art v 3 and Pru p 3 as a consequence of irreversible lanthionine formation.
Mol.Immunol., 116, 2019
|
|
3FQ8
 
 | | M248I mutant of GSAM | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Glutamate-1-semialdehyde 2,1-aminomutase | | Authors: | Stetefeld, J. | | Deposit date: | 2009-01-07 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Absence of a catalytic water confers resistance to the neurotoxin gabaculine.
Faseb J., 24, 2010
|
|
4EPP
 
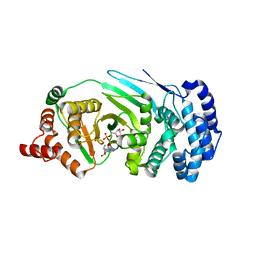 | |
4EPQ
 
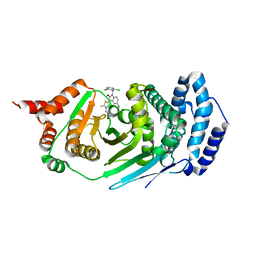 | |
3V2N
 
 | | COMPcc in complex with fatty acids | | Descriptor: | Cartilage Oligomerization matrix protein (coiled-coil domain), MYRISTIC ACID | | Authors: | Stetefeld, J. | | Deposit date: | 2011-12-12 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The pentameric channel of COMPcc in complex with different fatty acids.
Plos One, 7, 2012
|
|
3V2Q
 
 | | COMPcc in complex with fatty acids | | Descriptor: | Cartilage Oligomerization matrix protein (coiled-coil domain), PALMITIC ACID | | Authors: | Stetefeld, J. | | Deposit date: | 2011-12-12 | | Release date: | 2013-01-16 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The pentameric channel of COMPcc in complex with different fatty acids.
Plos One, 7, 2012
|
|
5NO5
 
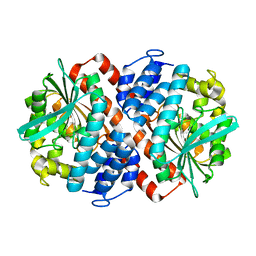 | | AbyA5 Wildtype | | Descriptor: | AbyA5 | | Authors: | Byrne, M.J, Race, P.R. | | Deposit date: | 2017-04-10 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Esterase-like Lyase Catalyzes Acetate Elimination in Spirotetronate/Spirotetramate Biosynthesis.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
7KLU
 
 | |
3SUX
 
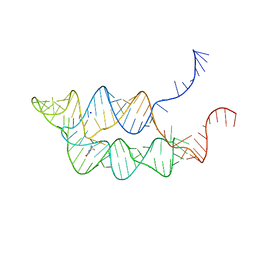 | | Crystal structure of THF riboswitch, bound with THF | | Descriptor: | 5-HYDROXYMETHYLENE-6-HYDROFOLIC ACID, Riboswitch, SODIUM ION | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Long-range pseudoknot interactions dictate the regulatory response in the tetrahydrofolate riboswitch.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3SUY
 
 | |
7W0X
 
 | |
7X5O
 
 | | Crystal structure of E. faecium SHMT in complex with Me-THF and PLP-Gly | | Descriptor: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, Serine hydroxymethyltransferase | | Authors: | Hasegawa, K, Hayashi, H. | | Deposit date: | 2022-03-05 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Serine hydroxymethyltransferase as a potential target of antibacterial agents acting synergistically with one-carbon metabolism-related inhibitors.
Commun Biol, 5, 2022
|
|
4DI4
 
 | | Crystal structure of a 3:1 complex of Treponema pallidum TatP(T) (Tp0957) bound to TatT (Tp0956) | | Descriptor: | TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, TatP(T) (Tp0957), ... | | Authors: | Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-01-30 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.714 Å) | | Cite: | Structural and Thermodynamic Characterization of the Interaction between Two Periplasmic Treponema pallidum Lipoproteins that are Components of a TPR-Protein-Associated TRAP Transporter (TPAT).
J.Mol.Biol., 420, 2012
|
|
1WSV
 
 | | Crystal Structure of Human T-protein of Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, SULFATE ION | | Authors: | Okamura-Ikeda, K, Hosaka, H, Yoshimura, M, Yamashita, E, Toma, S, Nakagawa, A, Fujiwara, K, Motokawa, Y, Taniguchi, H. | | Deposit date: | 2004-11-11 | | Release date: | 2005-08-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Human T-protein of Glycine Cleavage System at 2.0A Resolution and its Implication for Understanding Non-ketotic Hyperglycinemia
J.Mol.Biol., 351, 2005
|
|
2R4G
 
 | |
1PLU
 
 | |
2B2A
 
 | |
1S9A
 
 | | Crystal Structure of 4-Chlorocatechol 1,2-dioxygenase from Rhodococcus opacus 1CP | | Descriptor: | (1-HEXADECANOYL-2-TETRADECANOYL-GLYCEROL-3-YL) PHOSPHONYL CHOLINE, BENZOIC ACID, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Solyanikova, I.P, Kolomytseva, M.P, Scozzafava, A, Golovleva, L.A, Briganti, F. | | Deposit date: | 2004-02-04 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of 4-chlorocatechol 1,2-dioxygenase from the chlorophenol-utilizing gram-positive Rhodococcus opacus 1CP.
J.Biol.Chem., 279, 2004
|
|
1FRF
 
 | | CRYSTAL STRUCTURE OF THE NI-FE HYDROGENASE FROM DESULFOVIBRIO FRUCTOSOVORANS | | Descriptor: | FE (III) ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Montet, Y, Volbeda, A, Piras, C, Hatchikian, E.C, Frey, M, Fontecilla, J.C. | | Deposit date: | 1998-07-21 | | Release date: | 1998-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 3Fe-4S] to [4Fe-4S] cluster conversion in Desulfovibrio fructosovorans [NiFe] hydrogenase by site-directed mutagenesis
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
