8CYO
 
 | | Nurr1 Covalently Bound to a Synthetic Ligand, 10.25, via a Disulfide Bond | | Descriptor: | 2-(3,4-dichlorophenoxy)-N-(2-sulfanylethyl)acetamide, CHLORIDE ION, Nuclear receptor subfamily 4 group A member 2 | | Authors: | Bruning, J.M, Liu, J, England, P.M. | | Deposit date: | 2022-05-24 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Nurr1 Covalently Bound to a Synthetic Ligand, 10.25, via a Disulfide Bond
To Be Published
|
|
8DKN
 
 | | PPARg bound to T0070907 and Co-R peptide | | Descriptor: | 2-chloro-5-nitro-N-(pyridin-4-yl)benzamide, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Nuclear receptor corepressor 1 peptide, ... | | Authors: | Larsen, N.A, Tsai, J. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
8DSZ
 
 | | PPARg bound to partial agonist H3B-487 | | Descriptor: | (2R)-2-{5-[(5-{[(1R)-1-(4-tert-butylphenyl)ethyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Larsen, N.A. | | Deposit date: | 2022-07-24 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
8B4A
 
 | | Nativ complex of PqsE and RhlR with autoinducer C4-HSL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, FE (III) ION, N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, ... | | Authors: | Borgert, S.R, Blankenfeldt, W. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
7AOA
 
 | | Structure of the extended MTA1/HDAC1/MBD2/RBBP4 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, Histone-binding protein RBBP4, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (19.4 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
3D0S
 
 | |
6QNX
 
 | | Structure of the SA2/SCC1/CTCF complex | | Descriptor: | Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog, Transcriptional repressor CTCF | | Authors: | Li, Y, Muir, K.W, Panne, D. | | Deposit date: | 2019-02-12 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for cohesin-CTCF-anchored loops.
Nature, 578, 2020
|
|
4UF5
 
 | | Crystal structure of UCH-L5 in complex with inhibitory fragment of INO80G | | Descriptor: | NUCLEAR FACTOR RELATED TO KAPPA-B-BINDING PROTEIN, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | Authors: | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | Deposit date: | 2014-12-23 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
4UF6
 
 | | UCH-L5 in complex with ubiquitin-propargyl bound to an activating fragment of INO80G | | Descriptor: | NUCLEAR FACTOR RELATED TO KAPPA-B-BINDING PROTEIN, POLYUBIQUITIN-B, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | Authors: | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | Deposit date: | 2014-12-23 | | Release date: | 2015-03-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.69 Å) | | Cite: | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
7ABG
 
 | | Human pre-Bact-1 spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Cell division cycle 5-like protein, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
8J6W
 
 | |
7R3H
 
 | |
7R3E
 
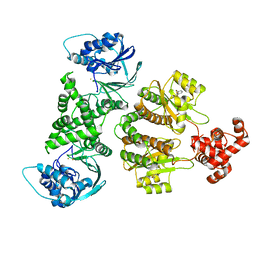 | | Fusion construct of PqsE and RhlR in complex with the synthetic antagonist mBTL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase,Regulatory protein RhlR, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, FE (III) ION | | Authors: | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
7R3J
 
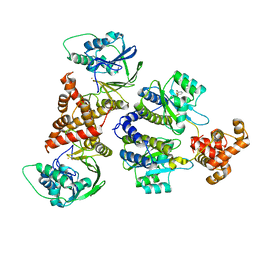 | | Nativ complex of PqsE and RhlR with the synthetic antagonist mBTL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, FE (III) ION, ... | | Authors: | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
7R3G
 
 | |
7R3I
 
 | |
6Y0G
 
 | | Structure of human ribosome in classical-PRE state | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Bhaskar, V, Schenk, A.D, Cavadini, S, von Loeffelholz, O, Natchiar, S.K, Klaholz, B.P, Chao, J.A. | | Deposit date: | 2020-02-07 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dynamics of uS19 C-Terminal Tail during the Translation Elongation Cycle in Human Ribosomes.
Cell Rep, 31, 2020
|
|
5NHK
 
 | | Structure of Ferric uptake regulator from francisella tularensis with Iron | | Descriptor: | FE (III) ION, Ferric uptake regulation protein, ZINC ION | | Authors: | Perard, J, Arnaud, L, Carpentier, P, Michaud-Soret, I, Cavazza, C. | | Deposit date: | 2017-03-21 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional studies of the metalloregulator Fur identify a promoter-binding mechanism and its role inFrancisella tularensisvirulence.
Commun Biol, 1, 2018
|
|
1Q07
 
 | | Crystal structure of the Au(I) form of E. coli CueR, a copper efflux regulator | | Descriptor: | GOLD ION, Transcriptional regulator cueR | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
5YZG
 
 | | The Cryo-EM Structure of Human Catalytic Step I Spliceosome (C complex) at 4.1 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Lei, J, Shi, Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-08-08 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of a human catalytic step I spliceosome
Science, 359, 2018
|
|
6Y2L
 
 | | Structure of human ribosome in POST state | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Bhaskar, V, Schenk, A.D, Cavadini, S, von Loeffelholz, O, Natchiar, S.K, Klaholz, B.P, Chao, J.A. | | Deposit date: | 2020-02-16 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Dynamics of uS19 C-Terminal Tail during the Translation Elongation Cycle in Human Ribosomes.
Cell Rep, 31, 2020
|
|
8FL4
 
 | |
8FL3
 
 | |
8FL2
 
 | |
6Y57
 
 | | Structure of human ribosome in hybrid-PRE state | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Bhaskar, V, Schenk, A.D, Cavadini, S, von Loeffelholz, O, Natchiar, S.K, Klaholz, B.P, Chao, J.A. | | Deposit date: | 2020-02-25 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Dynamics of uS19 C-Terminal Tail during the Translation Elongation Cycle in Human Ribosomes.
Cell Rep, 31, 2020
|
|
