3L1S
 
 | |
1Q6Q
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound xylitol 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-XYLITOL 5-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-08-13 | | Release date: | 2003-10-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Structural Evidence for a 1,2-Enediolate Intermediate in the Reaction Catalyzed by 3-Keto-l-Gulonate 6-Phosphate Decarboxylase, a Member of the Orotidine 5'-Monophosphate Decarboxylase Suprafamily
Biochemistry, 42, 2003
|
|
3EP5
 
 | | Human AdoMetDC E178Q mutant with no putrescine bound | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYRUVIC ACID, S-adenosylmethionine decarboxylase alpha chain, ... | | Authors: | Bale, S, Lopez, M.M, Makhatadze, G.I, Fang, Q, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2008-09-29 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis for Putrescine Activation of Human S-Adenosylmethionine Decarboxylase.
Biochemistry, 47, 2008
|
|
4CDW
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP4 | | Descriptor: | 1-[(3S)-5-(4-iodanylphenoxy)-3-methyl-pentyl]-3-pyridin-4-yl-imidazolidin-2-one, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-06 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
1XM2
 
 | | Crystal structure of Human PRL-1 | | Descriptor: | SULFATE ION, Tyrosine Phosphatase | | Authors: | Jeong, D.G, Kim, S.J, Kim, J.H, Son, J.H, Ryu, S.E. | | Deposit date: | 2004-10-01 | | Release date: | 2005-01-25 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Trimeric structure of PRL-1 phosphatase reveals an active enzyme conformation and regulation mechanisms
J.Mol.Biol., 345, 2005
|
|
4BY6
 
 | | Yeast Not1-Not2-Not5 complex | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bhaskar, V, Basquin, J, Conti, E. | | Deposit date: | 2013-07-18 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Structure and RNA-Binding Properties of the not1-not2-not5 Module of the Yeast Ccr4-not Complex
Nat.Struct.Mol.Biol., 20, 2013
|
|
1QCW
 
 | | Flavocytochrome B2, ARG289LYS mutant | | Descriptor: | FLAVOCYTOCHROME B2, N-SULFO-FLAVIN MONONUCLEOTIDE | | Authors: | Mowat, C.G, Durley, R.C.E, Pike, A.D, Barton, J.D, Chen, Z.-W, Mathews, F.S, Lederer, F, Reid, G.A, Chapman, S.K. | | Deposit date: | 1999-05-07 | | Release date: | 1999-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Kinetic and crystallographic studies on the active site Arg289Lys mutant of flavocytochrome b2 (yeast L-lactate dehydrogenase)
Biochemistry, 39, 2000
|
|
2OMX
 
 | | Crystal structure of InlA S192N G194S+S/hEC1 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epithelial-cadherin, ... | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamically reengineering the listerial invasion complex InlA/E-cadherin.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4B2P
 
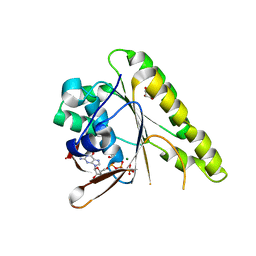 | | RadA C-terminal ATPase domain from Pyrococcus furiosus bound to GTP | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Marsh, M.E, Ehebauer, M.T, Scott, D, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2012-07-17 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | ATP Half-Sites in Rada and Rad51 Recombinases Bind Nucleotides
FEBS Open Bio, 6, 2016
|
|
7MDI
 
 | | Structure of the Neisseria gonorrhoeae ribonucleotide reductase in the inactive state | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Levitz, T.S, Drennan, C.L, Brignole, E.J. | | Deposit date: | 2021-04-05 | | Release date: | 2022-01-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Effects of chameleon dispense-to-plunge speed on particle concentration, complex formation, and final resolution: A case study using the Neisseria gonorrhoeae ribonucleotide reductase inactive complex.
J.Struct.Biol., 214, 2021
|
|
1FXV
 
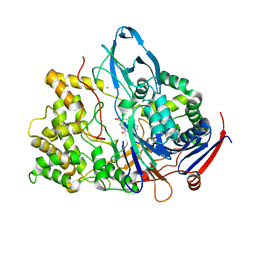 | | PENICILLIN ACYLASE MUTANT IMPAIRED IN CATALYSIS WITH PENICILLIN G IN THE ACTIVE SITE | | Descriptor: | CALCIUM ION, PENICILLIN ACYLASE, PENICILLIN G | | Authors: | Alkema, W.B, Hensgens, C.M, Kroezinga, E.H, de Vries, E, Floris, R, van der Laan, J.M, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2000-09-27 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and site-directed mutagenesis studies.
Protein Eng., 13, 2000
|
|
2PHB
 
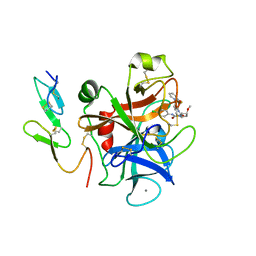 | | An Orally Efficacious Factor Xa Inhibitor | | Descriptor: | (2R,4R)-N~1~-(4-CHLOROPHENYL)-N~2~-[2-FLUORO-4-(2-OXOPYRIDIN-1(2H)-YL)PHENYL]-4-METHOXYPYRROLIDINE-1,2-DICARBOXAMIDE, CALCIUM ION, Coagulation factor X, ... | | Authors: | Zhang, E, Kohrt, J.T, Bigge, C.F, Finzel, B.C. | | Deposit date: | 2007-04-10 | | Release date: | 2008-03-25 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The discovery of (2R,4R)-N-(4-chlorophenyl)-N- (2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl)-4-methoxypyrrolidine-1,2-dicarboxamide (PD 0348292), an orally efficacious factor Xa inhibitor
Chem.Biol.Drug Des., 70, 2007
|
|
1XOC
 
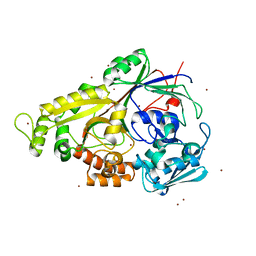 | | The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide. | | Descriptor: | Nonapeptide VDSKNTSSW, Oligopeptide-binding protein appA, ZINC ION | | Authors: | Levdikov, V.M, Blagova, E.V, Brannigan, J.A, Wright, L, Vagin, A.A, Wilkinson, A.J. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide.
J.Mol.Biol., 345, 2005
|
|
1UXV
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1ZRB
 
 | | Thrombin in complex with an azafluorenyl inhibitor 23b | | Descriptor: | 11-peptide hirudin fragment, 3-AZA-9-HYDROXY-9-FLUORENYLCARBONYL-L-PROLYL-2-AMINOMETHYL-5-CHLOROBENZYLAMIDE, N-OXIDE, ... | | Authors: | Stauffer, K.J, Williams, P.D, Selnick, H.G, Nantermet, P.G, Newton, C.L, Homnick, C.F, Zrada, M.M, Lewis, S.D, Lucas, B.J, Krueger, J.A, Pietrak, B.L, Lyle, E.A, Singh, R, Miller-Stein, C, White, R.B, Wong, B, Wallace, A.A, Sitko, G.R, Cook, J.J, Holahan, M.A, Stranieri-Michener, M, Leonard, Y.M, Lynch Jr, J.J, McMasters, D.R, Yan, Y. | | Deposit date: | 2005-05-19 | | Release date: | 2005-06-07 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 9-hydroxyazafluorenes and their use in thrombin inhibitors
J.Med.Chem., 48, 2005
|
|
5Z7R
 
 | |
4BA9
 
 | | The structural basis for the coordination of Y-family Translesion DNA Polymerases by Rev1 | | Descriptor: | DNA POLYMERASE KAPPA, DNA REPAIR PROTEIN REV1, MAGNESIUM ION, ... | | Authors: | Grummitt, C.G, Kilkenny, M.L, Frey, A, Roe, S.M, Oliver, A.W, Sale, J.E, Pearl, L.H. | | Deposit date: | 2012-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The Structural Basis for the Coordination of Y- Family Translesion DNA Polymerases by Rev1
To be Published
|
|
2Q2Q
 
 | | Structure of D-3-Hydroxybutyrate Dehydrogenase from Pseudomonas putida | | Descriptor: | Beta-D-hydroxybutyrate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Paithankar, K.S, Feller, C, Kuettner, E.B, Keim, A, Grunow, M, Strater, N. | | Deposit date: | 2007-05-29 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Cosubstrate-induced dynamics of D-3-hydroxybutyrate dehydrogenase from Pseudomonas putida.
Febs J., 274, 2007
|
|
3KZ4
 
 | | Crystal Structure of the Rotavirus Double Layered Particle | | Descriptor: | Inner capsid protein VP2, Intermediate capsid protein VP6, ZINC ION | | Authors: | Mcclain, B, Settembre, E.C, Bellamy, A.R, Harrison, S.C. | | Deposit date: | 2009-12-07 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | X-ray crystal structure of the rotavirus inner capsid particle at 3.8 A resolution.
J.Mol.Biol., 397, 2010
|
|
5ZA2
 
 | | Fox-4 beta-lactamase complexed with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase, ... | | Authors: | Nukaga, M, Hoshino, T, Papp-Wallace, K.M, Bonomo, R.A. | | Deposit date: | 2018-02-06 | | Release date: | 2018-03-07 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Probing the Mechanism of Inactivation of the FOX-4 Cephamycinase by Avibactam
Antimicrob. Agents Chemother., 62, 2018
|
|
2Q4Y
 
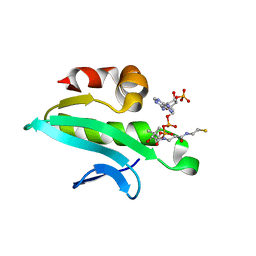 | | Ensemble refinement of the protein crystal structure of At1g77540-coenzyme A complex | | Descriptor: | COENZYME A, Uncharacterized protein At1g77540 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
3KIK
 
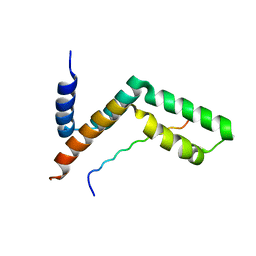 | | Sgf11:Sus1 complex | | Descriptor: | Protein SUS1, SAGA-associated factor 11 | | Authors: | Stewart, M, Ellisdon, A.M. | | Deposit date: | 2009-11-02 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the interaction between yeast Spt-Ada-Gcn5 acetyltransferase (SAGA) complex components Sgf11 and Sus1
J.Biol.Chem., 285, 2010
|
|
1PZK
 
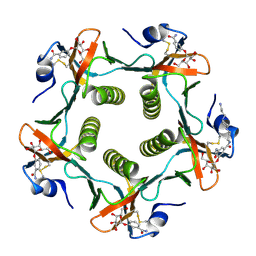 | | Cholera Toxin B-Pentamer Complexed With N-Acyl Phenyl Galactoside 9h | | Descriptor: | Cholera Toxin B Subunit, N-{3-[4-(3-AMINO-PROPYL)-PIPERAZIN-1-YL]-PROPYL}-3-(2-THIOPHEN-2-YL-ACETYLAMINO)-5-(3,4,5-TRIHYDROXY-6-HYDROXYMETHYL-TETRAHYDRO-PYRAN-2-YLOXY)-BENZAMIDE | | Authors: | Mitchell, D.D, Pickens, J.C, Korotkov, K, Fan, E, Hol, W.G.J. | | Deposit date: | 2003-07-11 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | 3,5-Substituted phenyl galactosides as leads in designing effective cholera toxin antagonists; synthesis and crystallographic studies
Bioorg.Med.Chem., 12, 2004
|
|
5TPX
 
 | | Bromodomain from Plasmodium Faciparum Gcn5, complexed with compound | | Descriptor: | (1S,2S)-N~1~,N~1~-dimethyl-N~2~-(3-methyl[1,2,4]triazolo[3,4-a]phthalazin-6-yl)-1-phenylpropane-1,2-diamine, CHLORIDE ION, Histone acetyltransferase GCN5, ... | | Authors: | Lin, Y.H, Hou, C.F.D, MOUSTAKIM, M, DIXON, D.J, Loppnau, P, Tempel, W, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, BRENNAN, P.E, Walker, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-10-21 | | Release date: | 2017-01-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a PCAF Bromodomain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4BGV
 
 | |
