1SNX
 
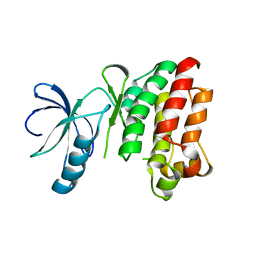 | | CRYSTAL STRUCTURE OF APO INTERLEUKIN-2 TYROSINE KINASE CATALYTIC DOMAIN | | Descriptor: | Tyrosine-protein kinase ITK/TSK | | Authors: | Brown, K, Long, J.M, Vial, S.C, Dedi, N, Dunster, N.J, Renwick, S.B, Tanner, A.J, Frantz, J.D, Fleming, M.A, Cheetham, G.M.T. | | Deposit date: | 2004-03-12 | | Release date: | 2004-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of interleukin-2 tyrosine kinase and their implications for the design of selective inhibitors.
J.Biol.Chem., 279, 2004
|
|
1SNY
 
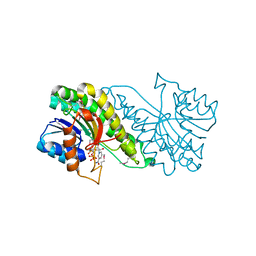 | | Carbonyl reductase Sniffer of D. melanogaster | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, sniffer CG10964-PA | | Authors: | Sgraja, T, Ulschmid, J, Becker, K, Schneuwly, S, Klebe, G, Reuter, K, Heine, A. | | Deposit date: | 2004-03-12 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights into the Neuroprotective-acting Carbonyl Reductase Sniffer of Drosophila melanogaster.
J.Mol.Biol., 342, 2004
|
|
1SNZ
 
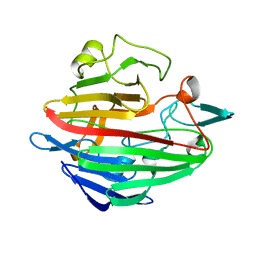 | |
1SO0
 
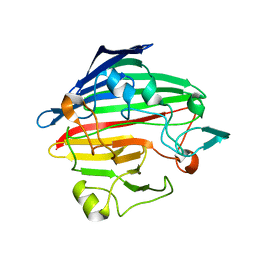 | | Crystal structure of human galactose mutarotase complexed with galactose | | Descriptor: | aldose 1-epimerase, beta-D-galactopyranose | | Authors: | Thoden, J.B, Timson, D.J, Reece, R.J, Holden, H.M. | | Deposit date: | 2004-03-12 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular structure of human galactose mutarotase
J.Biol.Chem., 279, 2004
|
|
1SO2
 
 | | CATALYTIC DOMAIN OF HUMAN PHOSPHODIESTERASE 3B In COMPLEX WITH A DIHYDROPYRIDAZINE INHIBITOR | | Descriptor: | 1-DEOXY-1-[(2-HYDROXYETHYL)(NONANOYL)AMINO]HEXITOL, 6-(4-{[2-(3-IODOBENZYL)-3-OXOCYCLOHEX-1-EN-1-YL]AMINO}PHENYL)-5-METHYL-4,5-DIHYDROPYRIDAZIN-3(2H)-ONE, MAGNESIUM ION, ... | | Authors: | Scapin, G, Patel, S.B, Chung, C, Varnerin, J.P, Edmondson, S.D, Mastracchio, A, Parmee, E.R, Becker, J.W, Singh, S.B, Van Der Ploeg, L.H, Tota, M.R. | | Deposit date: | 2004-03-12 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Human Phosphodiesterase 3B: Atomic Basis for Substrate and Inhibitor Specificity
Biochemistry, 43, 2004
|
|
1SO3
 
 | | Crystal structure of H136A mutant of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-03-12 | | Release date: | 2004-06-08 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Crystallographic Evidence for a Proton Relay System in the Active Site of 3-Keto-l-gulonate 6-Phosphate Decarboxylase(,)
Biochemistry, 43, 2004
|
|
1SO4
 
 | | Crystal structure of K64A mutant of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-03-12 | | Release date: | 2004-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Crystallographic Evidence for a Proton Relay System in the Active Site of 3-Keto-l-gulonate 6-Phosphate Decarboxylase(,)
Biochemistry, 43, 2004
|
|
1SO5
 
 | | Crystal structure of E112Q mutant of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-03-12 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Crystallographic Evidence for a Proton Relay System in the Active Site of 3-Keto-l-gulonate 6-Phosphate Decarboxylase(,)
Biochemistry, 43, 2004
|
|
1SO6
 
 | | Crystal structure of E112Q/H136A double mutant of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-03-12 | | Release date: | 2004-06-08 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Crystallographic Evidence for a Proton Relay System in the Active Site of 3-Keto-l-gulonate 6-Phosphate Decarboxylase(,)
Biochemistry, 43, 2004
|
|
1SO7
 
 | | Maltose-induced structure of the human cytolsolic sialidase Neu2 | | Descriptor: | CHLORIDE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Kato, R, Monti, E, Wakatsuki, S. | | Deposit date: | 2004-03-12 | | Release date: | 2004-11-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Structure of the Human Cytosolic Sialidase Neu2: EVIDENCE FOR THE DYNAMIC NATURE OF SUBSTRATE RECOGNITION
J.Biol.Chem., 280, 2005
|
|
1SO8
 
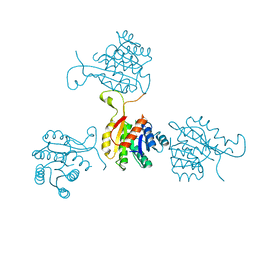 | | Abeta-bound human ABAD structure [also known as 3-hydroxyacyl-CoA dehydrogenase type II (Type II HADH), Endoplasmic reticulum-associated amyloid beta-peptide binding protein (ERAB)] | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase type II, CHLORIDE ION, SODIUM ION | | Authors: | Lustbader, J.W, Cirilli, M, Wu, H. | | Deposit date: | 2004-03-13 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ABAD directly links Abeta to mitochondrial toxicity in Alzheimer's disease.
Science, 304, 2004
|
|
1SO9
 
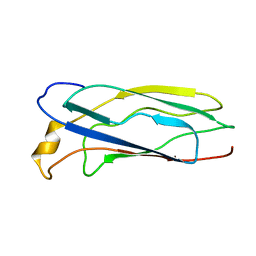 | | Solution Structure of apoCox11, 30 structures | | Descriptor: | Cytochrome C oxidase assembly protein ctaG | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
1SOA
 
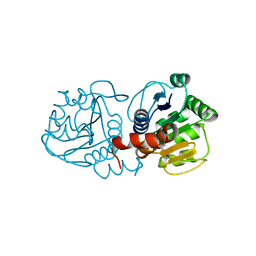 | | Human DJ-1 with sulfinic acid | | Descriptor: | RNA-binding protein regulatory subunit; oncogene DJ1 | | Authors: | Canet-Aviles, R, Wilson, M.A, Miller, D.W, Ahmad, R, McLendon, C, Bandyopadhyay, S, Baptista, M.J, Ringe, D, Petsko, G.A, Cookson, M.R. | | Deposit date: | 2004-03-13 | | Release date: | 2004-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1SOC
 
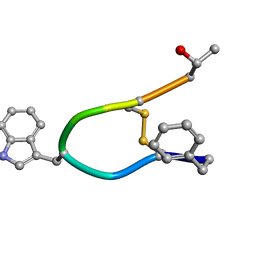 | |
1SOF
 
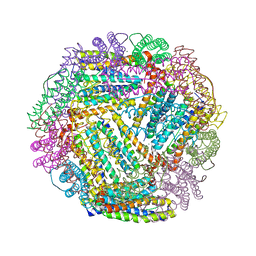 | | Crystal structure of the azotobacter vinelandii bacterioferritin at 2.6 A resolution | | Descriptor: | BARIUM ION, Bacterioferritin, FE (II) ION, ... | | Authors: | Liu, H.L, Huang, J.F, Bi, R.C. | | Deposit date: | 2004-03-14 | | Release date: | 2005-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 A resolution crystal structure of the bacterioferritin from Azotobacter vinelandii
Febs Lett., 573, 2004
|
|
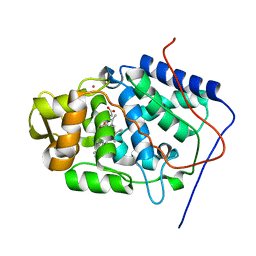
1SOG
 
 | |
1SOH
 
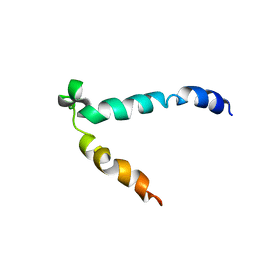 | |
1SOI
 
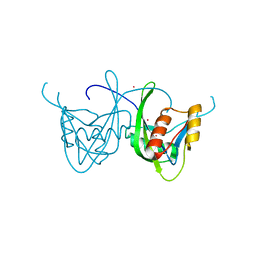 | | CRYSTAL STRUCTURE OF NUDIX HYDROLASE DR1025 IN COMPLEX WITH SM+3 | | Descriptor: | MutT/nudix family protein, SAMARIUM (III) ION | | Authors: | Ranatunga, W, Hill, E.E, Mooster, J.L, Holbrook, E.L, Schulze-Gahmen, U, Xu, W, Bessman, M.J, Brenner, S.E, Holbrook, S.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-15 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies of the Nudix Hydrolase DR1025 From Deinococcus radiodurans and its Ligand Complexes.
J.Mol.Biol., 339, 2004
|
|
1SOJ
 
 | | CATALYTIC DOMAIN OF HUMAN PHOSPHODIESTERASE 3B IN COMPLEX WITH IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, MAGNESIUM ION, cGMP-inhibited 3',5'-cyclic phosphodiesterase B | | Authors: | Scapin, G, Patel, S.B, Chung, C, Varnerin, J.P, Edmondson, S.D, Mastracchio, A, Parmee, E.R, Becker, J.W, Singh, S.B, Van Der Ploeg, L.H, Tota, M.R. | | Deposit date: | 2004-03-15 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Human Phosphodiesterase 3B: Atomic Basis for Substrate and Inhibitor Specificity
Biochemistry, 43, 2004
|
|
1SOK
 
 | | Crystal structure of the transthyretin mutant A108Y/L110E solved in space group p21212 | | Descriptor: | Transthyretin | | Authors: | Hornberg, A, Olofsson, A, Eneqvist, T, Lundgren, E, Sauer-Eriksson, A.E. | | Deposit date: | 2004-03-15 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The beta-strand D of transthyretin trapped in two discrete conformations
Biochim.Biophys.Acta, 1700, 2004
|
|
1SOL
 
 | | A PIP2 AND F-ACTIN-BINDING SITE OF GELSOLIN, RESIDUE 150-169 (NMR, AVERAGED STRUCTURE) | | Descriptor: | GELSOLIN (150-169) | | Authors: | Xian, W, Vegners, R, Janmey, P.A, Braunlin, W.H. | | Deposit date: | 1995-09-29 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Spectroscopic studies of a phosphoinositide-binding peptide from gelsolin: behavior in solutions of mixed solvent and anionic micelles.
Biophys.J., 69, 1995
|
|
1SOM
 
 | | TORPEDO CALIFORNICA ACETYLCHOLINESTERASE INHIBITED BY NERVE AGENT GD (SOMAN). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, METHYLPHOSPHONIC ACID ESTER GROUP, PROTEIN (ACETYLCHOLINESTERASE), ... | | Authors: | Greenblatt, H.M, Millard, C.B, Sussman, J.L, Silman, I. | | Deposit date: | 1999-03-17 | | Release date: | 1999-06-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of aged phosphonylated acetylcholinesterase: nerve agent reaction products at the atomic level.
Biochemistry, 38, 1999
|
|
1SON
 
 | | ADENYLOSUCCINATE SYNTHETASE IN COMPLEX WITH THE NATURAL FEEDBACK INHIBITOR AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENYLOSUCCINATE SYNTHETASE, BETA-MERCAPTOETHANOL | | Authors: | Cowan-Jacob, S.W. | | Deposit date: | 1996-05-07 | | Release date: | 1997-09-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The mode of action and the structure of a herbicide in complex with its target: binding of activated hydantocidin to the feedback regulation site of adenylosuccinate synthetase.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1SOO
 
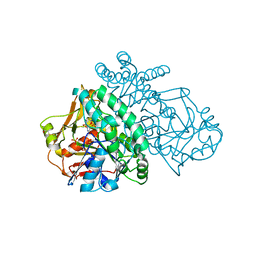 | | ADENYLOSUCCINATE SYNTHETASE INHIBITED BY HYDANTOCIDIN 5'-MONOPHOSPHATE | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE, BETA-MERCAPTOETHANOL, HYDANTOCIDIN-5'-PHOSPHATE, ... | | Authors: | Cowan-Jacob, S.W. | | Deposit date: | 1996-05-07 | | Release date: | 1997-09-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The mode of action and the structure of a herbicide in complex with its target: binding of activated hydantocidin to the feedback regulation site of adenylosuccinate synthetase.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1SOP
 
 | |
