2V5N
 
 | | STRUCTURE OF HUMAN IGF2R DOMAINS 11-12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Brown, J, Delaine, C, Zaccheo, O.J, Siebold, C, Gilbert, R.J, van Boxel, G, Denley, A, Wallace, J.C, Hassan, A.B, Forbes, B.E, Jones, E.Y. | | Deposit date: | 2007-07-06 | | Release date: | 2007-12-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Functional Analysis of the Igf-II/Igf2R Interaction
Embo J., 27, 2008
|
|
8E08
 
 | | Crystal structure of HPSE P6 in complex with tetraose pentosan inhibitor | | Descriptor: | 2,3,4-tri-O-sulfo-beta-D-xylopyranose-(1-4)-2,3-di-O-sulfo-beta-D-xylopyranose-(1-4)-2,3-di-O-sulfo-beta-D-xylopyranose-(1-4)-2,3-di-O-sulfo-beta-D-xylopyranose, ACETATE ION, Heparanase 50 kDa subunit, ... | | Authors: | Whitefield, C, Jackson, C.J. | | Deposit date: | 2022-08-08 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Complex Inhibitory Mechanism of Glycomimetics with Heparanase.
Biochemistry, 62, 2023
|
|
8GGT
 
 | | Locally refined cryoEM structure of receptor from beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #12 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8BOY
 
 | | X-ray structure of the adduct formed upon reaction of the five-coordinate Pt(II) complex, 1-Me,Me, with HEWL at pH 4.0 | | Descriptor: | 1-[1,3-dimethyl-4-(1~{H}-1,2,3-triazol-5-yl)imidazol-1-ium-2-yl]-1,2',11'-trimethyl-spiro[1$l^{6}-platinacycloprop-2-ene-1,15'-1,12-diaza-15$l^{6}-platinatetracyclo[10.2.1.0^{5,14}.0^{8,13}]pentadeca-2,4,6,8,10,13-hexaene], Lysozyme C, NITRATE ION, ... | | Authors: | Ferraro, G, Tito, G, Merlino, A. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Impact of Hydrophobic Chains in Five-Coordinate Glucoconjugate Pt(II) Anticancer Agents.
Int J Mol Sci, 24, 2023
|
|
2VFU
 
 | |
8GGH
 
 | | Structure of Trypanosoma (MDH)4-PEX5, distal conformation | | Descriptor: | Peroxisome targeting signal 1 receptor, malate dehydrogenase | | Authors: | Sonani, R.R, Artur, B, Jemiola-Rzeminska, M, Lipinski, O, Patel, S.N, Sood, T, Dubin, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Noncanonical interactions and conformational dynamics in cargo-Pex5-Pex14 ternary complex for peroxisomal import
Biorxiv, 2023
|
|
6SET
 
 | | X-ray structure of the gold/lysozyme adduct formed upon 3 days exposure of protein crystals to compound 1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ferraro, G, Giorgio, A, Merlino, A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein-mediated disproportionation of Au(i): insights from the structures of adducts of Au(iii) compounds bearing N,N-pyridylbenzimidazole derivatives with lysozyme.
Dalton Trans, 48, 2019
|
|
6VDI
 
 | |
8GG5
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #11 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
7LXD
 
 | | Structure of yeast DNA Polymerase Zeta (apo) | | Descriptor: | DNA polymerase delta small subunit, DNA polymerase delta subunit 3, DNA polymerase zeta catalytic subunit, ... | | Authors: | Truong, C.D, Craig, T.A, Cui, G, Botuyan, M.V, Serkasevich, R.A, Chan, K.-Y, Mer, G, Chiu, P.-L, Kumar, R. | | Deposit date: | 2021-03-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Cryo-EM reveals conformational flexibility in apo DNA polymerase zeta.
J.Biol.Chem., 297, 2021
|
|
3RFG
 
 | |
8GGP
 
 | | Locally refined cryoEM structure of receptor from beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #8 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GFZ
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #5 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
3KWA
 
 | | Polyamines inhibit carbonic anhydrases | | Descriptor: | Carbonic anhydrase 2, MERCURY (II) ION, SPERMINE, ... | | Authors: | Temperini, C. | | Deposit date: | 2009-12-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Polyamines inhibit carbonic anhydrases by anchoring to the zinc-coordinated water molecule
J.Med.Chem., 53, 2010
|
|
8GH1
 
 | | Locally refined cryoEM structure of receptor from beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #20 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GH2
 
 | | Structure of Trypanosoma (MDH)4-(Pex5)2, close conformation | | Descriptor: | Peroxisome targeting signal 1 receptor, malate dehydrogenase | | Authors: | Sonani, R.R, Artur, B, Jemiola-Rzeminska, M, Lipinski, O, Patel, S.N, Sood, T, Dubin, G. | | Deposit date: | 2023-03-09 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Noncanonical interactions and conformational dynamics in cargo-Pex5-Pex14 ternary complex for peroxisomal import
Biorxiv, 2023
|
|
8GGO
 
 | | Locally refined cryoEM structure of receptor from beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #7 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GG8
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #14 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GGQ
 
 | | Locally refined cryoEM structure of receptor from beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #9 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
5FKY
 
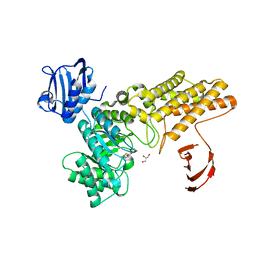 | | Structure of a hydrolase bound with an inhibitor | | Descriptor: | (3aR,5R,6S,7R,7aR)-2-amino-5-(hydroxymethyl)-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, GLYCEROL, O-GLCNACASE BT_4395 | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
5IXA
 
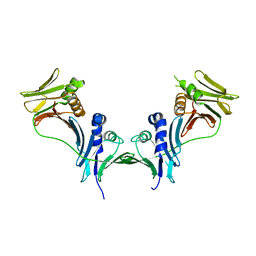 | |
8GG9
 
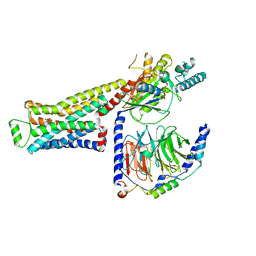 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #15 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GGR
 
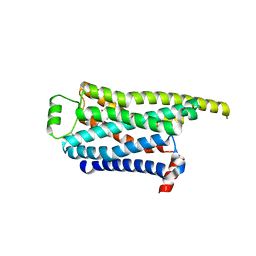 | | Locally refined cryoEM structure of receptor from beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #10 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
6T0G
 
 | | Crystal structure of CYP124 in complex with vitamin D3 | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, MAGNESIUM ION, Methyl-branched lipid omega-hydroxylase, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
5XOY
 
 | | Crystal structure of LysK from Thermus thermophilus in complex with Lysine | | Descriptor: | LYSINE, SULFATE ION, [LysW]-lysine hydrolase | | Authors: | Tomita, T, Fujita, S, Hasebe, F, Cho, S.-H, Yoshida, A, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2017-05-31 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of LysK, an enzyme catalyzing the last step of lysine biosynthesis in Thermus thermophilus, in complex with lysine: Insight into the mechanism for recognition of the amino-group carrier protein, LysW
Biochem. Biophys. Res. Commun., 491, 2017
|
|
