4L1N
 
 | |
4RDM
 
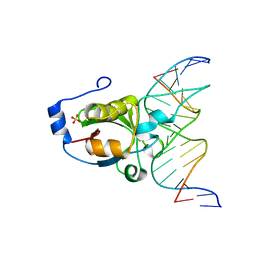 | | Crystal structure of R.NgoAVII restriction endonuclease B3 domain with cognate DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*AP*AP*GP*CP*GP*GP*CP*AP*AP*TP*CP*C)-3)', DNA (5'-D(*GP*GP*GP*AP*TP*TP*GP*CP*CP*GP*CP*TP*TP*AP*G)-3)', Restriction endonuclease R.NgoVII, ... | | Authors: | Tamulaitiene, G, Silanskas, A, Grazulis, S, Zaremba, M, Siksnys, V. | | Deposit date: | 2014-09-19 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the R-protein of the multisubunit ATP-dependent restriction endonuclease NgoAVII.
Nucleic Acids Res., 42, 2014
|
|
4R2H
 
 | |
3B2Y
 
 | |
4RHA
 
 | | Structure of the C-terminal domain of outer-membrane protein OmpA from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Outer membrane protein A, ... | | Authors: | Cuff, M.E, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-01 | | Release date: | 2014-10-29 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into PG-binding, conformational change, and dimerization of the OmpA C-terminal domains from Salmonella enterica serovar Typhimurium and Borrelia burgdorferi.
Protein Sci., 26, 2017
|
|
3B9T
 
 | |
3BB5
 
 | |
3BDD
 
 | |
3B5E
 
 | |
4JX2
 
 | |
4LII
 
 | |
4LG3
 
 | |
4LGC
 
 | |
3B5P
 
 | |
4L7A
 
 | |
3B8L
 
 | |
3BF5
 
 | |
3BDI
 
 | |
3BHN
 
 | |
4RJF
 
 | |
4LDA
 
 | |
4RWV
 
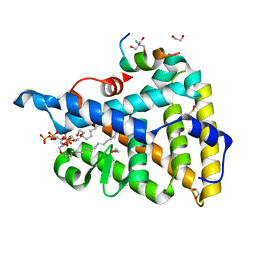 | | Crystal structure of PIP3 bound human nuclear receptor LRH-1 (Liver Receptor Homolog 1, NR5A2) in complex with a co-regulator DAX-1 (NR0B1) peptide at 1.86 A resolution | | Descriptor: | (2S)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dihexadecanoate, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell Biology, Partnership for Stem Cell Biology (STEMCELL) | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Crystal structure of a Homo sapiens hepatocytic transcription factor hB1F-2 (B1F2) in complex with nuclear receptor subfamily 0 group B member 1 (NR0B1, residues 140-154) from human at 1.86 A resolution
To be published
|
|
4RX6
 
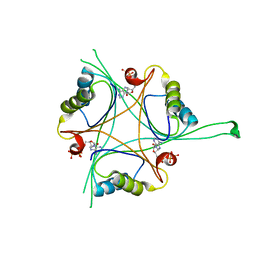 | | Structure of B. subtilis GlnK-ATP complex to 2.6 Angstrom | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Nitrogen regulatory PII-like protein | | Authors: | Schumacher, M.A, Cuthbert, B, Tonthat, N, Chinnam, N.G, Whitfill, T. | | Deposit date: | 2014-12-09 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5994 Å) | | Cite: | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
4K05
 
 | |
4QHW
 
 | |
