2DOO
 
 | | The structure of IMP-1 complexed with the detecting reagent (DansylC4SH) by a fluorescent probe | | Descriptor: | BETA-LACTAMASE IMP-1, N-[4-({[5-(DIMETHYLAMINO)-1-NAPHTHYL]SULFONYL}AMINO)BUTYL]-3-SULFANYLPROPANAMIDE, ZINC ION | | Authors: | Kurosaki, H, Yamaguchi, Y, Yasuzawa, H, Jin, W, Yamagata, Y, Arakawa, Y. | | Deposit date: | 2006-05-01 | | Release date: | 2006-11-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Probing, inhibition, and crystallographic characterization of metallo-beta-lactamase (IMP-1) with fluorescent agents containing dansyl and thiol groups
Chemmedchem, 1, 2006
|
|
1W22
 
 | | Crystal structure of inhibited human HDAC8 | | Descriptor: | HISTONE DEACETYLASE 8, N-HYDROXY-4-(METHYL{[5-(2-PYRIDINYL)-2-THIENYL]SULFONYL}AMINO)BENZAMIDE, POTASSIUM ION, ... | | Authors: | Vannini, A, Volpari, C, Caroli Casavola, E, Di Marco, S. | | Deposit date: | 2004-06-25 | | Release date: | 2004-09-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Eukaryotic Zn-Dependent Histone Deacetylase,Human Hdac8,Complexed with a Hydroxamic Acid Inhibitor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1U5U
 
 | |
1WB0
 
 | | specificity and affinity of natural product cyclopentapeptide inhibitor Argifin against human chitinase | | Descriptor: | ARGIFIN, CHITOTRIOSIDASE 1, GLYCEROL, ... | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Hodkinson, M, Adams, D.J, Shiomi, K, Omura, S, Van Aalten, D.M.F. | | Deposit date: | 2004-10-29 | | Release date: | 2005-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Specificity and Affinity of Natural Product Cyclopentapeptide Inhibitors Against Aspergillus Fumigatus, Human and Bacterial Chitinases
Chem.Biol., 12, 2005
|
|
2DQ0
 
 | | Crystal structure of seryl-tRNA synthetase from Pyrococcus horikoshii complexed with a seryl-adenylate analog | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, SULFATE ION, Seryl-tRNA synthetase | | Authors: | Itoh, Y, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-18 | | Release date: | 2007-06-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic and mutational studies of seryl-tRNA synthetase from the archaeon Pyrococcus horikoshii.
Rna Biol., 5, 2008
|
|
1WAC
 
 | | Back-priming mode of Phi6 RNA-dependent RNA polymerase | | Descriptor: | P2 PROTEIN | | Authors: | Laurila, M.R.L, Salgado, P.S, Stuart, D.I, Grimes, J.M, Bamford, D.H. | | Deposit date: | 2004-10-26 | | Release date: | 2005-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Back-Priming Mode of Phi6 RNA-Dependent RNA Polymerase
J.Gen.Virol., 86, 2005
|
|
1UCB
 
 | |
1W68
 
 | | Crystal Structure of Mouse Ribonucleotide Reductase Subunit R2 under Oxidizing Conditions. A Fully Occupied Dinuclear Iron Cluster. | | Descriptor: | MU-OXO-DIIRON, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE M2 CHAIN | | Authors: | Karlsen, S, Strand, K.R, Kolberg, M, Rohr, A.K, Gorbitz, C.H, Andersson, K.K. | | Deposit date: | 2004-08-16 | | Release date: | 2004-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structural Studies of Changes in the Native Dinuclear Iron Center of Ribonucleotide Reductase Protein R2 from Mouse
J.Biol.Chem., 279, 2004
|
|
2DQT
 
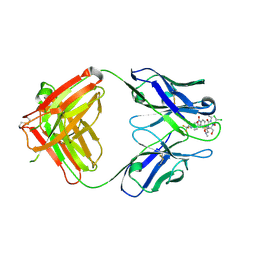 | | High resolution crystal structure of the complex of the hydrolytic antibody Fab 6D9 and a transition-state analog | | Descriptor: | IMMUNOGLOBULIN 6D9, [1-(3-DIMETHYLAMINO-PROPYL)-3-ETHYL-UREIDO]-[4-(2,2,2-TRIFLUORO-ACETYLAMINO)-BENZYL]PHOSPHINIC ACID-2-(2,2-DIHYDRO-ACETYLAMINO)-3-HYDROXY-1-(4-NITROPHENYL)-PROPYL ESTER | | Authors: | Kristensen, O, Vassylyev, D.G, Tanaka, F, Ito, N, Morikawa, K, Fujii, I. | | Deposit date: | 2006-05-30 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
2DL2
 
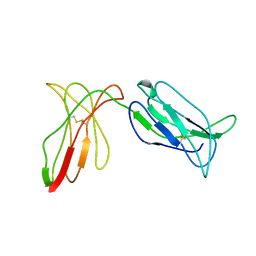 | | KILLER IMMUNOGLOBULIN RECEPTOR 2DL2 | | Descriptor: | PROTEIN (MHC CLASS I NK CELL RECEPTOR PRECURSOR (P58 NATURAL KILLER CELL RECEPTOR CLONE CL-43)) | | Authors: | Sun, P, Snyder, G. | | Deposit date: | 1999-03-08 | | Release date: | 1999-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the HLA-Cw3 allotype-specific killer cell inhibitory receptor KIR2DL2
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1UDQ
 
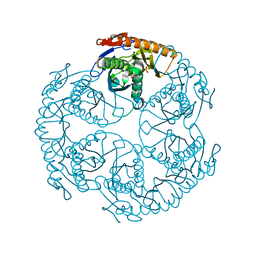 | | Crystal structure of the tRNA processing enzyme RNase PH T125A mutant from Aquifex aeolicus | | Descriptor: | PHOSPHATE ION, Ribonuclease PH, SULFATE ION | | Authors: | Ishii, R, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-02 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the tRNA Processing Enzyme RNase PH from Aquifex aeolicus
J.Biol.Chem., 278, 2003
|
|
2DR7
 
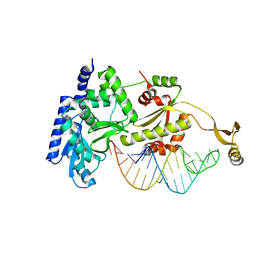 | | Complex structure of CCA-adding enzyme with tRNAminiDC | | Descriptor: | CCA-adding enzyme, SULFATE ION, tRNA (33-MER) | | Authors: | Tomita, K, Ishitani, R, Fukai, S, Nureki, O. | | Deposit date: | 2006-06-08 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complete crystallographic analysis of the dynamics of CCA sequence addition
Nature, 443, 2006
|
|
2DRI
 
 | |
2DRU
 
 | | Crystal structure and binding properties of the CD2 and CD244 (2B4) binding protein, CD48 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, chimera of CD48 antigen and T-cell surface antigen CD2 | | Authors: | Evans, E.J, Ikemizu, S, Davis, S.J. | | Deposit date: | 2006-06-15 | | Release date: | 2006-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and Binding Properties of the CD2 and CD244 (2B4)-binding Protein, CD48
J.Biol.Chem., 281, 2006
|
|
1W9P
 
 | | Specificity and affinity of natural product cyclopentapeptide inhibitors against Aspergillus fumigatus, human and bacterial chitinaseFra | | Descriptor: | CHITINASE, SULFATE ION | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Hodkinson, M, Adams, D.J, Shiomi, K, Omura, S, Van Aalten, D.M.F. | | Deposit date: | 2004-10-15 | | Release date: | 2004-10-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Specificity and Affinity of Natural Product Cyclopentapeptide Inhibitors Against Aspergillus Fumigatus, Human and Bacterial Chitinases
Chem.Biol., 12, 2005
|
|
1U7B
 
 | |
1UEU
 
 | |
2DSI
 
 | | Crystal structure of Glu171 to Arg mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Saraboji, K, Malathy Sony, S.M, Ponnuswamy, M.N, Kumarevel, T.S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-06-30 | | Release date: | 2006-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
1VIE
 
 | |
2DTM
 
 | | Thermodynamic and structural analyses of hydrolytic mechanism by catalytic antibodies | | Descriptor: | IMMUNOGLOBULIN 6D9 | | Authors: | Oda, M, Ito, N, Tsumuraya, T, Suzuki, K, Fujii, I. | | Deposit date: | 2006-07-13 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
1U8E
 
 | | HUMAN DIPEPTIDYL PEPTIDASE IV/CD26 MUTANT Y547F | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bjelke, J.R, Christensen, J, Branner, S, Wagtmann, N, Olsen, C, Kanstrup, A.B, Rasmussen, H.B. | | Deposit date: | 2004-08-05 | | Release date: | 2004-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tyrosine 547 constitutes an essential part of the catalytic mechanism of dipeptidyl peptidase IV
J.Biol.Chem., 279, 2004
|
|
1VCP
 
 | |
1VIN
 
 | |
1VUB
 
 | | CCDB, A TOPOISOMERASE POISON FROM E. COLI | | Descriptor: | CCDB, CHLORIDE ION | | Authors: | Loris, R, Dao-Thi, M.-H, Bahasi, E.M, Van Melderen, L, Poortmans, F, Liddington, R, Couturier, M, Wyns, L. | | Deposit date: | 1998-04-17 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of CcdB, a topoisomerase poison from E. coli.
J.Mol.Biol., 285, 1999
|
|
1VFR
 
 | | THE MAJOR NAD(P)H:FMN OXIDOREDUCTASE FROM VIBRIO FISCHERI | | Descriptor: | FLAVIN MONONUCLEOTIDE, NAD(P)H:FMN OXIDOREDUCTASE | | Authors: | Koike, H, Sasaki, H, Kobori, T, Zenno, S, Saigo, K, Murphy, M.E.P, Adman, E.T, Tanokura, M. | | Deposit date: | 1998-01-09 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 A crystal structure of the major NAD(P)H:FMN oxidoreductase of a bioluminescent bacterium, Vibrio fischeri: overall structure, cofactor and substrate-analog binding, and comparison with related flavoproteins.
J.Mol.Biol., 280, 1998
|
|
