2BLH
 
 | | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nienhaus, K, Ostermann, A, Nienhaus, G.U, Parak, F.G, Schmidt, M. | | Deposit date: | 2005-03-04 | | Release date: | 2005-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W
Biochemistry, 44, 2005
|
|
2BJS
 
 | | Isopenicillin N synthase C-terminal truncation mutant | | Descriptor: | FE (III) ION, ISOPENICILLIN N SYNTHETASE, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, ... | | Authors: | McNeill, L.A, Sami, M, Clifton, I.J, Burzlaff, N.I. | | Deposit date: | 2005-02-07 | | Release date: | 2006-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Terminally Truncated Isopenicillin N Synthase Generates a Dithioester Product: Evidence for a Thioaldehyde Intermediate during Catalysis and a New Mode of Reaction for Non-Heme Iron Oxidases.
Chemistry, 23, 2017
|
|
2BT2
 
 | | Structure of the regulator of G-protein signaling 16 | | Descriptor: | REGULATOR OF G-PROTEIN SIGNALING 16 | | Authors: | Bunkoczi, G, Haroniti, A, Longman, E, Niesen, F, Soundararajan, M, Ball, L.J, von Delft, F, Doyle, D.A, Arrowsmith, C, Edwards, A, Sundstrom, M. | | Deposit date: | 2005-05-25 | | Release date: | 2005-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Diversity in the Rgs Domain and its Interaction with Heterotrimeric G Protein Alpha- Subunits.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2BQ6
 
 | | Crystal structure of factor Xa in complex with 21 | | Descriptor: | 1-{[5-(5-CHLORO-2-THIENYL)ISOXAZOL-3-YL]METHYL}-3-CYANO-N-(1-ISOPROPYLPIPERIDIN-4-YL)-7-METHYL-1H-INDOLE-2-CARBOXAMIDE, CALCIUM ION, COAGULATION FACTOR X, ... | | Authors: | Nazare, M, Will, D.W, Matter, H, Schreuder, H, Ritter, K, Urmann, M, Essrich, M, Bauer, A, Wagner, M, Czech, J, Laux, V, Wehner, V. | | Deposit date: | 2005-04-27 | | Release date: | 2006-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the Subpockets of Factor Xa Reveals Two Binding Modes for Inhibitors Based on a 2-Carboxyindole Scaffold: A Study Combining Structure-Activity Relationship and X-Ray Crystallography.
J.Med.Chem., 48, 2005
|
|
2BGV
 
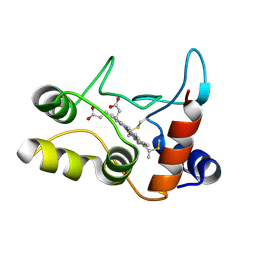 | | X-ray structure of ferric cytochrome c-550 from Paracoccus versutus | | Descriptor: | CYTOCHROME C-550, HEME C | | Authors: | Worrall, J.A.R, Van Roon, A.-M.M, Ubbink, M, Canters, G.W. | | Deposit date: | 2005-01-05 | | Release date: | 2005-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Effect of Replacing the Axial Methionine Ligand with a Lysine Residue in Cytochrome C-550 from Paracoccus Versutus Assessed by X-Ray Crystallography and Unfolding.
FEBS J., 272, 2005
|
|
2BNX
 
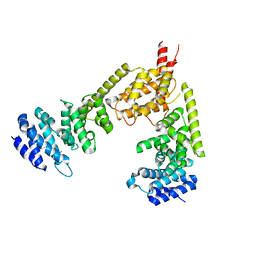 | | Crystal structure of the dimeric regulatory domain of mouse diaphaneous-related formin (DRF), mDia1 | | Descriptor: | CHLORIDE ION, DIAPHANOUS PROTEIN HOMOLOG 1 | | Authors: | Otomo, T, Otomo, C, Tomchick, D.R, Machius, M, Rosen, M.K. | | Deposit date: | 2005-04-05 | | Release date: | 2005-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Rho Gtpase-Mediated Activation of the Formin Mdia1
Mol.Cell, 18, 2005
|
|
2C7N
 
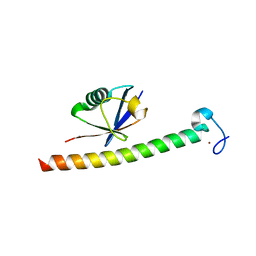 | | Human Rabex-5 residues 1-74 in complex with Ubiquitin | | Descriptor: | RAB GUANINE NUCLEOTIDE EXCHANGE FACTOR 1, UBIQUITIN, ZINC ION | | Authors: | Penengo, L, Mapelli, M, Murachelli, A.G, Confalioneri, S, Magri, L, Musacchio, A, Di Fiore, P.P, Polo, S, Schneider, T.R. | | Deposit date: | 2005-11-25 | | Release date: | 2006-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Ubiquitin Binding Domains of Rabex-5 Reveals Two Modes of Interaction with Ubiquitin.
Cell(Cambridge,Mass.), 124, 2006
|
|
2CII
 
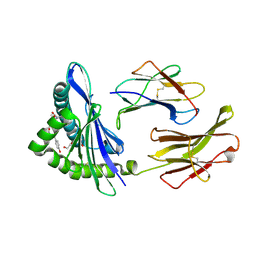 | | The crystal structure of H-2Db complexed with a partial peptide epitope suggests an MHC Class I assembly-intermediate | | Descriptor: | BETA-2-MICROGLOBULIN, GLYCEROL, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN D-B ALPHA CHAIN, ... | | Authors: | Glithero, A, Tormo, J, Doering, K, Kojima, M, Jones, E.Y, Elliott, T. | | Deposit date: | 2006-03-21 | | Release date: | 2006-03-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of H-2D(b) complexed with a partial peptide epitope suggests a major histocompatibility complex class I assembly intermediate.
J. Biol. Chem., 281, 2006
|
|
2C7C
 
 | | FITTED COORDINATES FOR GROEL-ATP7-GROES CRYO-EM COMPLEX (EMD-1180) | | Descriptor: | 10 KDA CHAPERONIN MOLECULE: GROES, PROTEIN CPN10, GROES PROTEIN, ... | | Authors: | Ranson, N.A, Clare, D.K, Farr, G.W, Houldershaw, D, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2005-11-22 | | Release date: | 2006-01-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Allosteric Signalling of ATP Hydrolysis in Groel-Groes Complexes.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2C9S
 
 | | 1.24 Angstroms resolution structure of Zn-Zn Human Superoxide dismutase | | Descriptor: | ACETATE ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Strange, R.W, Antonyuk, S.V, Hough, M.A, Doucette, P.A, Valentine S, J.S, Hasnain, S. | | Deposit date: | 2005-12-14 | | Release date: | 2005-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Variable Metallation of Human Superoxide Dismutase: Atomic Resolution Crystal Structures of Cu-Zn, Zn-Zn and as-Isolated Wild-Type Enzymes.
J.Mol.Biol., 356, 2006
|
|
2CXF
 
 | | RUN domain of Rap2 interacting protein x, crystallized in C2 space group | | Descriptor: | rap2 interacting protein x | | Authors: | Kukimoto-Niino, M, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-29 | | Release date: | 2005-12-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Crystal Structure of the RUN Domain of the RAP2-interacting Protein x
J.Biol.Chem., 281, 2006
|
|
2C7M
 
 | | Human Rabex-5 residues 1-74 in complex with Ubiquitin | | Descriptor: | RAB GUANINE NUCLEOTIDE EXCHANGE FACTOR 1, UBIQUITIN, ZINC ION | | Authors: | Penengo, L, Mapelli, M, Murachelli, A.G, Confalioneri, S, Magri, L, Musacchio, A, Di Fiore, P.P, Polo, S, Schneider, T.R. | | Deposit date: | 2005-11-25 | | Release date: | 2006-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin.
Cell, 124, 2006
|
|
2C9U
 
 | | 1.24 Angstroms resolution structure of as-isolated Cu-Zn Human Superoxide dismutase | | Descriptor: | ACETATE ION, COPPER (II) ION, SULFATE ION, ... | | Authors: | Strange, R.W, Antonyuk, S.V, Hough, M.A, Doucette, P.A, Valentine S, J.S, Hasnain, S. | | Deposit date: | 2005-12-14 | | Release date: | 2005-12-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Variable Metallation of Human Superoxide Dismutase: Atomic Resolution Crystal Structures of Cu-Zn, Zn-Zn and as-Isolated Wild-Type Enzymes.
J.Mol.Biol., 356, 2006
|
|
2ATB
 
 | | Triple mutant 8D9D10V of scorpion toxin LQH-alpha-IT | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NITRATE ION, ... | | Authors: | Kahn, R, Karbat, I, Gurevitz, M, Frolow, F. | | Deposit date: | 2005-08-24 | | Release date: | 2006-09-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structures of Lqh-alpha-IT and Lqh-alpha-IT8D9D10V mutant
To be Published
|
|
2AMN
 
 | | Solution structure of Fowlicidin-1, a novel Cathelicidin antimicrobial peptide from chicken | | Descriptor: | cathelicidin | | Authors: | Xiao, Y, Dai, H, Bommineni, Y.R, Prakash, O, Zhang, G. | | Deposit date: | 2005-08-09 | | Release date: | 2006-07-18 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure-activity relationships of fowlicidin-1, a cathelicidin antimicrobial peptide in chicken.
Febs J., 273, 2006
|
|
2BH4
 
 | | X-ray structure of the M100K variant of ferric cyt c-550 from Paracoccus versutus determined at 100 K. | | Descriptor: | CYTOCHROME C-550, HEME C | | Authors: | Worrall, J.A.R, Van Roon, A.-M.M, Ubbink, M, Canters, G.W. | | Deposit date: | 2005-01-07 | | Release date: | 2005-05-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Effect of Replacing the Axial Methionine Ligand with a Lysine Residue in Cytochrome C-550 from Paracoccus Versutus Assessed by X-Ray Crystallography and Unfolding.
FEBS J., 272, 2005
|
|
2BLY
 
 | | HEWL after a high dose x-ray "burn" | | Descriptor: | LYSOZYME C, TETRAETHYLENE GLYCOL | | Authors: | Nanao, M.H, Ravelli, R.B. | | Deposit date: | 2005-03-08 | | Release date: | 2005-09-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Improving Radiation-Damage Substructures for Rip.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BF6
 
 | |
2EEX
 
 | | Crystal structure of Cel44A, GH family 44 endoglucanase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitago, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2007-02-19 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
2ECP
 
 | | THE CRYSTAL STRUCTURE OF THE E. COLI MALTODEXTRIN PHOSPHORYLASE COMPLEX | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLYCEROL, MALTODEXTRIN PHOSPHORYLASE, ... | | Authors: | O'Reilly, M, Watson, K.A, Johnson, L.N. | | Deposit date: | 1998-10-27 | | Release date: | 1999-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of the Escherichia coli maltodextrin phosphorylase-acarbose complex.
Biochemistry, 38, 1999
|
|
2EFW
 
 | | Crystal structure of the RTP:nRB complex from Bacillus subtilis | | Descriptor: | DNA (5'-D(*DCP*DT*DAP*DTP*DGP*DTP*DAP*DCP*DCP*DAP*DAP*DAP*DTP*DGP*DTP*DTP*DCP*DAP*DGP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DCP*DTP*DGP*DAP*DAP*DCP*DAP*DTP*DTP*DTP*DGP*DGP*DTP*DAP*DCP*DAP*DTP*DAP*DG)-3'), Replication termination protein | | Authors: | Vivian, J.P, Porter, C.J, Wilce, J.A, Wilce, M.C.J. | | Deposit date: | 2007-02-26 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An asymmetric structure of the Bacillus subtilis replication terminator protein in complex with DNA
J.Mol.Biol., 370, 2007
|
|
2B4J
 
 | | Structural basis for the recognition between HIV-1 integrase and LEDGF/p75 | | Descriptor: | GLYCEROL, Integrase (IN), PC4 and SFRS1 interacting protein, ... | | Authors: | Cherepanov, P, Ambrosio, A.L, Rahman, S, Ellenberger, T, Engelman, A. | | Deposit date: | 2005-09-24 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B9K
 
 | |
2ASC
 
 | | Scorpion toxin LQH-alpha-IT | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ETHANOL, ... | | Authors: | Kahn, R, Karbat, I, Gurevitz, M, Frolow, F. | | Deposit date: | 2005-08-23 | | Release date: | 2006-09-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray structures of Lqh-alpha-IT and Lqh-alpha-IT8D9D10V mutant
To be Published
|
|
2AGH
 
 | | Structural basis for cooperative transcription factor binding to the CBP coactivator | | Descriptor: | Crebbp protein, Myb proto-oncogene protein, Zinc finger protein HRX | | Authors: | De Guzman, R.N, Goto, N.K, Dyson, H.J, Wright, P.E. | | Deposit date: | 2005-07-26 | | Release date: | 2005-11-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Cooperative Transcription Factor Binding to the CBP Coactivator
J.Mol.Biol., 355, 2006
|
|
