5FT0
 
 | | Crystal structure of gp37(Dip) from bacteriophage phiKZ | | Descriptor: | ARGININE, GP37, POTASSIUM ION | | Authors: | Van den Bossche, A, Hardwick, S.W, Ceyssens, P.J, Hendrix, H, Voet, M, Dendooven, T, Bandyra, K.J, De Maeyer, M, Aertsen, A, Noben, J.P, Luisi, B.F, Lavigne, R. | | Deposit date: | 2016-01-08 | | Release date: | 2016-08-03 | | Last modified: | 2017-03-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural elucidation of a novel mechanism for the bacteriophage-based inhibition of the RNA degradosome.
Elife, 5, 2016
|
|
6Y7I
 
 | | Structure of the BRD9 bromodomain and compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 1-(8-pyridin-2-ylpyrrolo[1,2-a]pyrimidin-6-yl)ethanone, Bromodomain-containing protein 9 | | Authors: | Diaz-Saez, L, Krojer, T, Picaud, S, von Delft, F, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A, Bountra, C, Huber, K.V.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-01 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the BRD9 bromodomain and compound 2
To Be Published
|
|
5FU3
 
 | | The complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition | | Descriptor: | CBM74-RFGH5, SODIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Basle, A, Luis, A.S, Venditto, I, Gilbert, H.J. | | Deposit date: | 2016-01-20 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8AZB
 
 | |
5VIG
 
 | | Crystal structure of anti-Zika antibody Z006 bound to Zika virus envelope protein DIII | | Descriptor: | CITRATE ANION, Fab heavy chain, Fab light chain, ... | | Authors: | Keeffe, J.R, West Jr, A.P, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-04-16 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico.
Cell, 169, 2017
|
|
7Q7R
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 1 | | Descriptor: | 2-chloranyl-4-[[(2S)-2-cyclopropyl-3,3-bis(fluoranyl)-7-methyl-6-oxidanylidene-2,4-dihydro-1H-[1,4]oxazepino[2,3-c]quinolin-10-yl]amino]pyridine-3-carbonitrile, B-cell lymphoma 6 protein, CHLORIDE ION | | Authors: | Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optimizing Shape Complementarity Enables the Discovery of Potent Tricyclic BCL6 Inhibitors.
J.Med.Chem., 65, 2022
|
|
5FIE
 
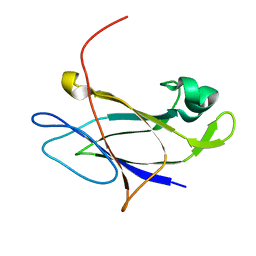 | | Crystal structure of N-terminal domain of shaft pilin spaA from Lactobacillus rhamnosus GG | | Descriptor: | Cell surface protein SpaA, SODIUM ION | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
6FQJ
 
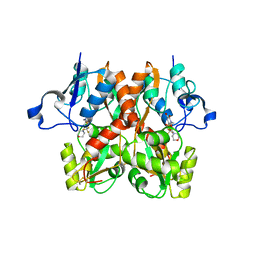 | | GluA2(flop) G724C ligand binding core dimer bound to ZK200775 at 2.50 Angstrom resolution | | Descriptor: | Glutamate receptor 2,Glutamate receptor 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.500017 Å) | | Cite: | Homomeric GluA2(R) AMPA receptors can conduct when desensitized.
Nat Commun, 10, 2019
|
|
6Y7J
 
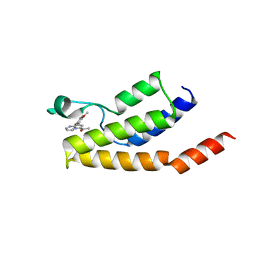 | | Structure of the BRD9 bromodomain and compound 15 | | Descriptor: | 1-[8-(2,5-dimethoxyphenyl)pyrrolo[1,2-a]pyrimidin-6-yl]ethanone, ACETATE ION, Bromodomain-containing protein 9, ... | | Authors: | Diaz-Saez, L, Krojer, T, Picaud, S, von Delft, F, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A, Bountra, C, Huber, K.V.M. | | Deposit date: | 2020-03-01 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the BRD9 bromodomain
To Be Published
|
|
6QA4
 
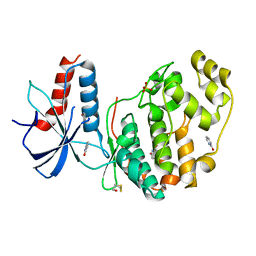 | | ERK2 mini-fragment binding | | Descriptor: | 1~{H}-pyridin-2-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
5FW1
 
 | | Crystal structure of SpyCas9 variant VQR bound to sgRNA and TGAG PAM target DNA | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Bargsten, K, Jinek, M. | | Deposit date: | 2016-02-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural Plasticity of Pam Recognition by Engineered Variants of the RNA-Guided Endonuclease Cas9.
Mol.Cell, 61, 2016
|
|
6Q3Y
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the inhibitor 16i | | Descriptor: | (7~{R})-2-[[2-ethoxy-4-(1-methylpiperidin-4-yl)phenyl]amino]-7-ethyl-5-methyl-8-(phenylmethyl)-7~{H}-pteridin-6-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Heidenreich, D, Watts, E, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S, Hoelder, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Designing Dual Inhibitors of Anaplastic Lymphoma Kinase (ALK) and Bromodomain-4 (BRD4) by Tuning Kinase Selectivity.
J.Med.Chem., 62, 2019
|
|
8AIC
 
 | |
6Y7K
 
 | | Structure of the BRD9 bromodomain and compound 27 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 9, SODIUM ION, ... | | Authors: | Diaz-Saez, L, Krojer, T, Picaud, S, von Delft, F, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A, Bountra, C, Huber, K.V.M. | | Deposit date: | 2020-03-01 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the BRD9 bromodomain
To Be Published
|
|
6Q42
 
 | | Crystal Structure of Human Pancreatic Phospholipase A2 | | Descriptor: | CHLORIDE ION, Phospholipase A2 | | Authors: | Saul, F, Haouz, A, Lambeau, G, Theze, J. | | Deposit date: | 2018-12-05 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PLA2G1B is involved in CD4 anergy and CD4 lymphopenia in HIV-infected patients.
J.Clin.Invest., 130, 2020
|
|
6YE9
 
 | | Small-molecule inhibitor of 14-3-3 protein-protein interactions | | Descriptor: | 14-3-3 protein sigma, MAGNESIUM ION, [2-[2-oxidanylidene-2-[(phenylmethyl)amino]ethoxy]phenyl]phosphonic acid | | Authors: | Ottmann, C, Visser, E.J. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based conversion of a promiscuous inhibitor to a selective stabilizer of protein-protein interactions
To Be Published
|
|
8AUG
 
 | |
6QA8
 
 | | Glycogen Phosphorylase b in complex with 28 | | Descriptor: | (5~{S},7~{R},8~{S},9~{S},10~{R})-7-(hydroxymethyl)-8,9,10-tris(oxidanyl)-2-phenyl-6-oxa-1,3-diazaspiro[4.5]dec-1-en-4-one, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Glucopyranosylidene-spiro-imidazolinones, a New Ring System: Synthesis and Evaluation as Glycogen Phosphorylase Inhibitors by Enzyme Kinetics and X-ray Crystallography.
J.Med.Chem., 62, 2019
|
|
6FWJ
 
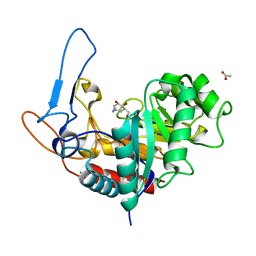 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-(1,2-anhydro-carba-mannosamine) and alpha-1,2-mannobiose | | Descriptor: | (1~{R},2~{R},3~{R},4~{R},6~{R})-4-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2018-03-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
6Q48
 
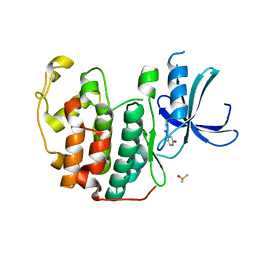 | | CDK2 in complex with FragLite7 | | Descriptor: | 4-iodanyl-3~{H}-pyridin-2-one, Cyclin-dependent kinase 2, DIMETHYL SULFOXIDE | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6Y4N
 
 | | Structure of Tubulin Tyrosine Ligase in Complex with Tb116 | | Descriptor: | (2~{R})-1-methylpiperidine-2-carboxylic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Gavrilyuk, J, Nocek, B, Rigol, S, Nicolaou, K.C, Stoll, V. | | Deposit date: | 2020-02-21 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Tubulysin Analogues, Linker-Drugs, and Antibody-Drug Conjugates, Insights into Structure-Activity Relationships, and Tubulysin-Tubulin Binding Derived from X-ray Crystallographic Analysis.
J.Org.Chem., 86, 2021
|
|
5FJ3
 
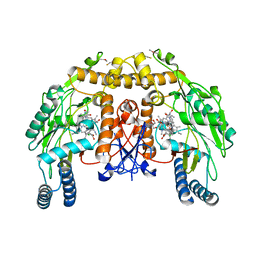 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 7-((4-CHLORO-3-((METHYLAMINO)METHYL) PHENOXY)METHYL)QUINOLIN-2-AMINE in the absence of acetate | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[4-chloranyl-3-(methylaminomethyl)phenoxy]methyl]quinolin-2-amine, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-10-06 | | Release date: | 2015-10-28 | | Last modified: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phenyl Ether- and Aniline-Containing 2-Aminoquinolines as Potent and Selective Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
7QPX
 
 | |
6FM2
 
 | | CARP domain of mouse cyclase-associated protein 1 (CAP1) bound to ADP-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Kotila, T.M, Kogan, K, Lappalainen, P. | | Deposit date: | 2018-01-30 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of actin monomer re-charging by cyclase-associated protein.
Nat Commun, 9, 2018
|
|
6Q4H
 
 | | CDK2 in complex with FragLite36 | | Descriptor: | 2-[3-[(2-azanyl-9~{H}-purin-6-yl)oxy]phenyl]ethanoic acid, Cyclin-dependent kinase 2, DIMETHYL SULFOXIDE | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
