7RJE
 
 | | Complex III2 from Candida albicans, Inz-5 bound | | Descriptor: | 3-[2-fluoro-5-(trifluoromethyl)phenyl]-7-methyl-1-[(2-methyl-2H-tetrazol-5-yl)methyl]-1H-indazole, Cytochrome b, Cytochrome b-c1 complex subunit 2, ... | | Authors: | Di Trani, J.M, Rubinstein, J.L. | | Deposit date: | 2021-07-20 | | Release date: | 2021-09-15 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Rieske head domain dynamics and indazole-derivative inhibition of Candida albicans complex III.
Structure, 30, 2022
|
|
1WDS
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose, ... | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
7RB7
 
 | | Room temperature structure of hAChE in complex with substrate analog 4K-TMA and MMB4 oxime | | Descriptor: | 1,1'-methylenebis{4-[(E)-(hydroxyimino)methyl]pyridin-1-ium}, 4,4-DIHYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, Acetylcholinesterase | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2021-07-05 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Room temperature crystallography of human acetylcholinesterase bound to a substrate analogue 4K-TMA: Towards a neutron structure
Curr Res Struct Biol, 3, 2021
|
|
7RB6
 
 | | Low temperature structure of hAChE in complex with substrate analog 4K-TMA | | Descriptor: | 4,4-DIHYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2021-07-05 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room temperature crystallography of human acetylcholinesterase bound to a substrate analogue 4K-TMA: Towards a neutron structure
Curr Res Struct Biol, 3, 2021
|
|
1WQV
 
 | | Human Factor Viia-Tissue Factor Complexed with propylsulfonamide-D-Thr-Met-p-aminobenzamidine | | Descriptor: | CALCIUM ION, Coagulation factor VII, N-[DIHYDROXY(PROPYL)-LAMBDA~4~-SULFANYL]THREONYL-N~1~-{4-[AMINO(IMINO)METHYL]BENZYL}METHIONINAMIDE, ... | | Authors: | Kadono, S, Sakamoto, A, Kikuchi, Y, Oh-eda, M, Yabuta, N, Koga, T, Hattori, K, Shiraishi, T, Haramura, M, Kodama, H. | | Deposit date: | 2004-10-02 | | Release date: | 2005-10-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human factor VIIa/tissue factor in complex with peptide mimetic inhibitor
Biochem.Biophys.Res.Commun., 324, 2004
|
|
7RB5
 
 | | Room temperature structure of hAChE in complex with substrate analog 4K-TMA | | Descriptor: | 4,4-DIHYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, Acetylcholinesterase | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2021-07-05 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Room temperature crystallography of human acetylcholinesterase bound to a substrate analogue 4K-TMA: Towards a neutron structure
Curr Res Struct Biol, 3, 2021
|
|
1P6J
 
 | | Rat neuronal NOS heme domain with L-N(omega)-nitroarginine-(4R)-amino-L-proline amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, L-N(OMEGA)-NITROARGININE-(4R)-AMINO-L-PROLINE AMIDE, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2HJI
 
 | | Structural model for the Fe-containing isoform of acireductone dioxygenase | | Descriptor: | E-2/E-2' protein, FE (II) ION | | Authors: | Pochapsky, T.C, Ju, T, Maroney, M.J, Chai, S.C. | | Deposit date: | 2006-06-30 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | One Protein, Two Enzymes Revisited: A Structural Entropy Switch Interconverts the Two Isoforms of Acireductone Dioxygenase
J.Mol.Biol., 363, 2006
|
|
2HTL
 
 | | Structure of the Escherichia coli ClC chloride channel Y445F mutant and Fab complex | | Descriptor: | BROMIDE ION, Fab fragment, Heavy chain, ... | | Authors: | Accardi, A, Lobet, S, Williams, C, Miller, C, Dutzler, R. | | Deposit date: | 2006-07-26 | | Release date: | 2006-09-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Synergism Between Halide Binding and Proton Transport in a CLC-type Exchanger.
J.Mol.Biol., 362, 2006
|
|
1WT1
 
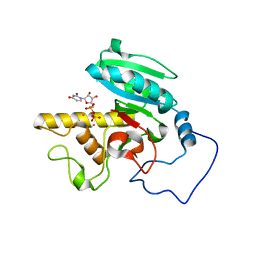 | | Mutant ABO(H) blood group glycosyltransferase with bound UDP and acceptor | | Descriptor: | Histo-blood group ABO system transferase, MERCURY (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Lee, H.J, Barry, C.H, Borisova, S.N, Seto, N.O.L, Zheng, R.B, Blancher, A, Evans, S.V, Palcic, M.M. | | Deposit date: | 2004-11-12 | | Release date: | 2004-12-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the inactivity of human blood group o2 glycosyltransferase
J.Biol.Chem., 280, 2005
|
|
2HKP
 
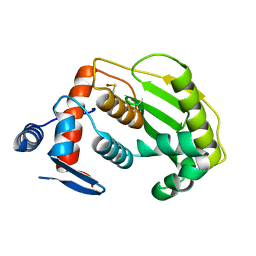 | |
2HNZ
 
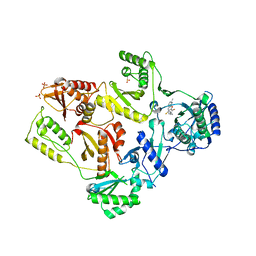 | | Crystal Structure of E138K Mutant HIV-1 Reverse Transcriptase in Complex with PETT-2 | | Descriptor: | 1-[2-(4-ETHOXY-3-FLUOROPYRIDIN-2-YL)ETHYL]-3-(5-METHYLPYRIDIN-2-YL)THIOUREA, PHOSPHATE ION, Reverse transcriptase/ribonuclease H | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-13 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
1WUA
 
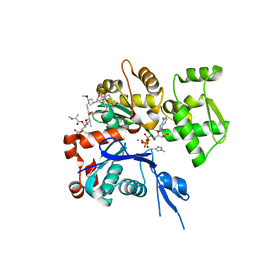 | | The structure of Aplyronine A-actin complex | | Descriptor: | (8R,9R,10R,11R,14S,18S,20S,24S)-24-{(1R,2S,3R,6R,7R,8R,9S,10E)-8-(ACETYLOXY)-6-[(N,N-DIMETHYLALANYL)OXY]-11-[FORMYL(MET HYL)AMINO]-2-HYDROXY-1,3,7,9-TETRAMETHYLUNDEC-10-ENYL}-10-HYDROXY-14,20-DIMETHOXY-9,11,15,18-TETRAMETHYL-2-OXOOXACYCLOTE TRACOSA-3,5,15,21-TETRAEN-8-YL N,N,O-TRIMETHYLSERINATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Hirata, K, Muraoka, S, Suenaga, K, Kuroda, T, Kato, K, Tanaka, H, Yamamoto, M, Takata, M, Yamada, K, Kigoshi, H. | | Deposit date: | 2004-12-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure basis for antitumor effect of aplyronine a
J.Mol.Biol., 356, 2006
|
|
2HMI
 
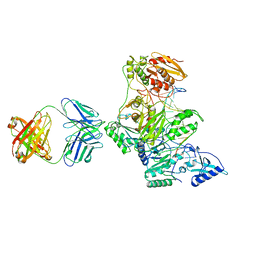 | | HIV-1 REVERSE TRANSCRIPTASE/FRAGMENT OF FAB 28/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3'), FAB FRAGMENT OF MONOCLONAL ANTIBODY 28, ... | | Authors: | Ding, J, Arnold, E. | | Deposit date: | 1998-04-10 | | Release date: | 1998-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 A resolution.
J.Mol.Biol., 284, 1998
|
|
2HNL
 
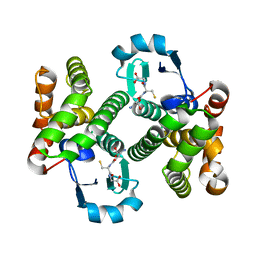 | | Structure of the prostaglandin D synthase from the parasitic nematode Onchocerca volvulus | | Descriptor: | GLUTATHIONE, Glutathione S-transferase 1 | | Authors: | Perbandt, M, Hoppner, J, Betzel, C, Liebau, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the extracellular glutathione S-transferase OvGST1 from the human pathogenic parasite Onchocerca volvulus.
J.Mol.Biol., 377, 2008
|
|
1OUQ
 
 | | Crystal structure of wild-type Cre recombinase-loxP synapse | | Descriptor: | Cre recombinase, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Ennifar, E, Meyer, J.E.W, Buchholz, F, Stewart, A.F, Suck, D. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a wild-type Cre recombinase-loxP synapse reveals a novel spacer conformation suggesting an alternative mechanism for DNA cleavage activation
Nucleic Acids Res., 31, 2003
|
|
1P6M
 
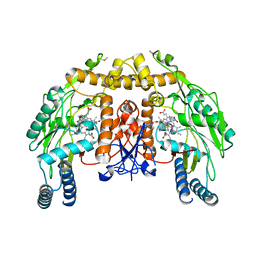 | | Bovine endothelial NOS heme domain with (4S)-N-(4-amino-5-[aminoethyl]aminopentyl)-N'-nitroguanidine bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1X0K
 
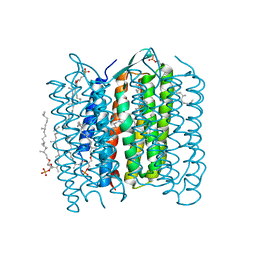 | | Crystal Structure of Bacteriorhodopsin at pH 10 | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Okumura, H, Murakami, M, Kouyama, T. | | Deposit date: | 2005-03-23 | | Release date: | 2005-08-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Acid Blue and Alkaline Purple Forms of Bacteriorhodopsin
J.Mol.Biol., 351, 2005
|
|
1X2A
 
 | | Crystal Structure of e.coli AspAT complexed with N-phosphopyridoxyl-D-glutamic acid | | Descriptor: | Aspartate aminotransferase, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-D-GLUTAMIC ACID | | Authors: | Goto, M. | | Deposit date: | 2005-04-21 | | Release date: | 2005-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of C5-dicarboxylic substrate to aspartate aminotransferase: implications for the conformational change at the transaldimination step.
Biochemistry, 44, 2005
|
|
2HTK
 
 | | Structure of the Escherichia coli ClC chloride channel Y445A mutant and Fab complex | | Descriptor: | BROMIDE ION, Fab fragment, Heavy chain, ... | | Authors: | Accardi, A, Lobet, S, Williams, C, Miller, C, Dutzler, R. | | Deposit date: | 2006-07-26 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Synergism between halide binding and proton transport in a CLC-type exchanger
J.Mol.Biol., 362, 2006
|
|
7RMA
 
 | | Structure of the fourth UIM (Ubiquitin Interacting Motif) of ANKRD13D in complex with a high affinity UbV (Ubiquitin Variant) | | Descriptor: | Ankyrin repeat domain-containing protein 13D, SODIUM ION, SULFATE ION, ... | | Authors: | Singer, A.U, Manczyk, N, Veggiani, G, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-07-27 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Panel of Engineered Ubiquitin Variants Targeting the Family of Human Ubiquitin Interacting Motifs.
Acs Chem.Biol., 17, 2022
|
|
2HPA
 
 | | STRUCTURAL ORIGINS OF L(+)-TARTRATE INHIBITION OF HUMAN PROSTATIC ACID PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-PROPYL-TARTRAMIC ACID, ... | | Authors: | Lacount, M.W, Handy, G, Lebioda, L. | | Deposit date: | 1998-09-11 | | Release date: | 1998-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural origins of L(+)-tartrate inhibition of human prostatic acid phosphatase.
J.Biol.Chem., 273, 1998
|
|
7RIB
 
 | | Griffithsin mutant Y28F/Y68F/Y110F | | Descriptor: | Griffithsin, SULFATE ION, alpha-D-mannopyranose | | Authors: | Zhao, G, Sun, J, Bewley, C.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
7RKI
 
 | | Griffithsin-S10Y/S42Y/S88Y | | Descriptor: | Griffithsin, alpha-D-mannopyranose | | Authors: | Sun, J.D, Zhao, G.X. | | Deposit date: | 2021-07-22 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
7RIA
 
 | | Griffithsin variant Y28A/Y68A/Y110A | | Descriptor: | Griffithsin, SULFATE ION, alpha-D-mannopyranose | | Authors: | Zhao, G, Sun, J, Bewley, C.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
