7T0U
 
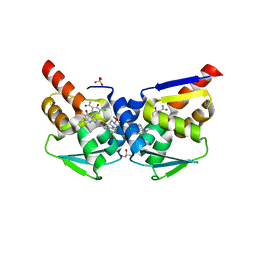 | | Crystal structure of the BCL6 BTB domain in complex with OICR-12387 | | Descriptor: | 3-chloro-5-{7-[2-({5-chloro-2-[(2S)-2-methyl-4-(oxetan-3-yl)piperazin-1-yl]pyridin-4-yl}amino)-2-oxoethyl]-3-methyl-4-oxo-2-(trifluoromethyl)-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl}-2-hydroxybenzamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-11-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of the BCL6 BTB domain in complex with OICR-12387
To Be Published
|
|
7T0T
 
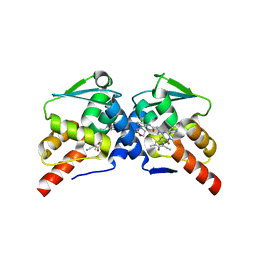 | | Crystal structure of the BCL6 BTB domain in complex with OICR-10562 | | Descriptor: | DIMETHYL SULFOXIDE, Isoform 2 of B-cell lymphoma 6 protein, N-[5-chloro-2-(morpholin-4-yl)pyridin-4-yl]-2-[5-(3-cyano-4-hydroxy-5-methylphenyl)-3-methyl-2-(1-methyl-1H-pyrazol-4-yl)-4-oxo-3,4-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl]acetamide | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-11-30 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the BCL6 BTB domain in complex with OICR-10562
To Be Published
|
|
7T0S
 
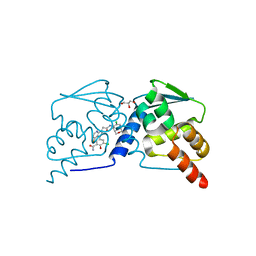 | | Crystal structure of the BCL6 BTB domain in complex with OICR-11864 | | Descriptor: | 3-chloro-5-{1-[2-({5-chloro-2-[(3S)-3-methylmorpholin-4-yl]pyridin-4-yl}amino)-2-oxoethyl]-4-fluoro-1H-pyrrolo[2,3-b]pyridin-3-yl}-2-hydroxybenzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-11-30 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the BCL6 BTB domain in complex with OICR-11864
To Be Published
|
|
6FTO
 
 | |
6GKO
 
 | | Mouse thymidylate synthase cocrystallized with dUMP and soaked in phenolphthalein | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 3,3-bis(4-hydroxyphenyl)-2-benzofuran-1-one, DIMETHYL SULFOXIDE, ... | | Authors: | Maj, P, Wilk, P, Jarmula, A, Weiss, M.S, Rode, W. | | Deposit date: | 2018-05-21 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | Thymidylate synthase fragment screening
To Be Published
|
|
6GYJ
 
 | | Mouse thymidylate synthase crystal soaked in phenolphthalein | | Descriptor: | 3,3-bis(4-hydroxyphenyl)-2-benzofuran-1-one, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Maj, P, Jarmula, A, Wilk, P, Weiss, M.S, Rode, W. | | Deposit date: | 2018-06-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thymidylate synthase fragment screening
To be published
|
|
3DVP
 
 | | Pak1 peptide bound LC8 | | Descriptor: | Dynein light chain 1, cytoplasmic, P21 activated Kinase peptide | | Authors: | Lightcap, C.M, Williams, J.C. | | Deposit date: | 2008-07-18 | | Release date: | 2009-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical and structural characterization of the Pak1-LC8 interaction.
J.Biol.Chem., 283, 2008
|
|
7ZZT
 
 | | Ligand binding to HDAC2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
7ZZW
 
 | | Ligand binding to HDAC2 | | Descriptor: | 1,2-ETHANEDIOL, 2-(cyclohexylazaniumyl)ethanesulfonate, CALCIUM ION, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
8EJV
 
 | | The crystal structure of Pseudomonas putida PcaR in complex with succinate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Pham, C, Skarina, T, Di Leo, R, Stogios, P.J, Mahadevan, R, Savchenko, A. | | Deposit date: | 2022-09-19 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The crystal structure of Pseudomonas putida PcaR in complex with succinate
To Be Published
|
|
8EJU
 
 | | The crystal structure of Pseudomonas putida PcaR | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Transcription regulatory protein (Pca regulon), ... | | Authors: | Pham, C, Skarina, T, Di Leo, R, Stogios, P.J, Mahadevan, R, Savchenko, A. | | Deposit date: | 2022-09-19 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The crystal structure of Pseudomonas putida PcaR
To Be Published
|
|
3CFS
 
 | | Structural basis of the interaction of RbAp46/RbAp48 with histone H4 | | Descriptor: | ARSENIC, Histone H4, Histone-binding protein RBBP7 | | Authors: | Murzina, N.V, Pei, X.-Y, Pratap, J.V, Sparkes, M, Vicente-Garcia, J, Ben-Shahar, T.R, Verreault, A, Luisi, B.F, Laue, E.D. | | Deposit date: | 2008-03-04 | | Release date: | 2008-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Recognition of Histone H4 by the Histone-Chaperone RbAp46.
Structure, 16, 2008
|
|
7ZZU
 
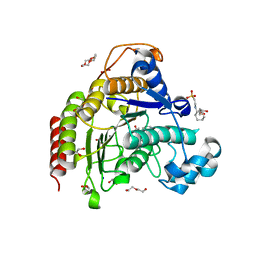 | | Inhibitory Ligand binding to HDAC2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[(2~{R},4~{S})-4-phenylpyrrolidin-2-yl]carbonylpiperazin-1-yl]pyridine-3-carbonitrile, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
8A0B
 
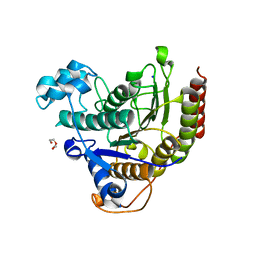 | | Inhibitor binding to HDAC2 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-dihydroisoindol-2-yl-[(2R,4S)-4-phenylpyrrolidin-1-ium-2-yl]methanone, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-27 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
2W99
 
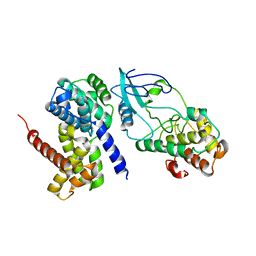 | | Crystal Structure of CDK4 in complex with a D-type cyclin | | Descriptor: | CELL DIVISION PROTEIN KINASE 4, G1/S-SPECIFIC CYCLIN-D1 | | Authors: | Day, P.J, Cleasby, A, Tickle, I.J, Reilly, M.O, Coyle, J.E, Holding, F.P, McMenamin, R.L, Yon, J, Chopra, R, Lengauer, C, Jhoti, H. | | Deposit date: | 2009-01-22 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human Cdk4 in Complex with a D-Type Cyclin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7BHY
 
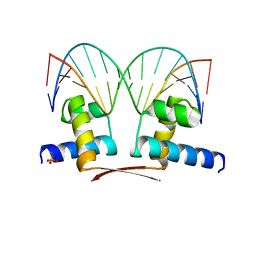 | | DNA-binding domain of DeoR in complex with the DNA operator | | Descriptor: | DNA operator - strand 1, DNA operator - strand 2, Deoxyribonucleoside regulator, ... | | Authors: | Novakova, M, Rezacova, P, Skerlova, J, Brynda, J. | | Deposit date: | 2021-01-11 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into DNA recognition by bacterial transcriptional regulators of the SorC/DeoR family.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7AOA
 
 | | Structure of the extended MTA1/HDAC1/MBD2/RBBP4 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, Histone-binding protein RBBP4, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (19.4 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
7M3X
 
 | | Crystal Structure of the Apo Form of Human RBBP7 | | Descriptor: | Histone-binding protein RBBP7, UNKNOWN ATOM OR ION | | Authors: | Righetto, G.L, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-19 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of the Apo Form of Human RBBP7
To Be Published
|
|
7AO9
 
 | | Structure of the core MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
7AO8
 
 | | Structure of the MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
7M4T
 
 | | Menin bound to M-1121 | | Descriptor: | Menin, methyl {(1S,2R)-2-[(1S)-2-(azetidin-1-yl)-1-(3-fluorophenyl)-1-{1-[(3-methoxy-1-{4-[(1S,4S)-5-propanoyl-2,5-diazabicyclo[2.2.1]heptane-2-sulfonyl]phenyl}azetidin-3-yl)methyl]piperidin-4-yl}ethyl]cyclopentyl}carbamate, praseodymium triacetate | | Authors: | Stuckey, J. | | Deposit date: | 2021-03-22 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Discovery of M-1121 as an Orally Active Covalent Inhibitor of Menin-MLL Interaction Capable of Achieving Complete and Long-Lasting Tumor Regression.
J.Med.Chem., 64, 2021
|
|
7COW
 
 | | 353 bp di-nucleosome harboring cohesive DNA termini with linker histone H1.0 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (353-MER), ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2020-08-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
3HUU
 
 | |
7N40
 
 | | Crystal structure of LIN9-RbAp48-LIN37, a MuvB subcomplex | | Descriptor: | Histone-binding protein RBBP4, Isoform 2 of Protein lin-9 homolog, Protein lin-37 homolog | | Authors: | Asthana, A, Ramanan, P, Tripathi, S.M, Rubin, S.M. | | Deposit date: | 2021-06-02 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The MuvB complex binds and stabilizes nucleosomes downstream of the transcription start site of cell-cycle dependent genes.
Nat Commun, 13, 2022
|
|
7LZR
 
 | |
