1HKN
 
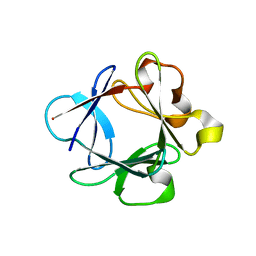 | | A complex between acidic fibroblast growth factor and 5-amino-2-naphthalenesulfonate | | Descriptor: | 5-AMINO-NAPHTALENE-2-MONOSULFONATE, HEPARIN-BINDING GROWTH FACTOR 1 | | Authors: | Fernandez-Tornero, C, Lozano, R.M, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2003-03-10 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Leads for Development of New Naphthalenesulfonate Derivatives with Enhanced Antiangiogenic Activity: Crystal Structure of Acidic Fibroblast Growth Factor in Complex with 5-Amino-2-Naphthalenesulfonate
J.Biol.Chem., 278, 2003
|
|
1HL3
 
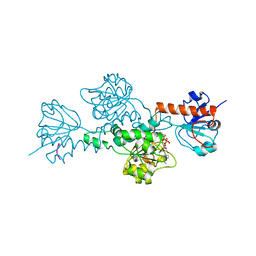 | | CtBP/BARS in ternary complex with NAD(H) and PIDLSKK peptide | | Descriptor: | C-TERMINAL BINDING PROTEIN 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PRO-ILE-ASP-LEU-SER-LYS-LYS PEPTIDE | | Authors: | Nardini, M, Spano, S, Cericola, C, Pesce, A, Massaro, A, Millo, E, Luini, A, Corda, D, Bolognesi, M. | | Deposit date: | 2003-03-13 | | Release date: | 2003-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Ctbp/Bars: A Dual-Function Protein Involved in Transcription Co-Repression and Golgi Membrane Fission
Embo J., 22, 2003
|
|
4FCQ
 
 | | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization | | Descriptor: | 4-(2,4-dimethylphenyl)-2-(methylsulfanyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Heat shock protein HSP 90-alpha | | Authors: | Davies, N.G, Browne, H, Davis, B, Foloppe, N, Geoffrey, S, Gibbons, B, Hart, T, Drysdale, M.J, Mansell, H, Massey, A, Matassova, N, Moore, J.D, Murray, J, Pratt, R, Ray, S, Roughley, S.D, Jensen, M.R, Schoepfer, J, Scriven, K, Simmonite, H, Stokes, S, Surgenor, A, Webb, P, Wright, L, Brough, P. | | Deposit date: | 2012-05-25 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization.
Bioorg.Med.Chem., 20, 2012
|
|
4XPB
 
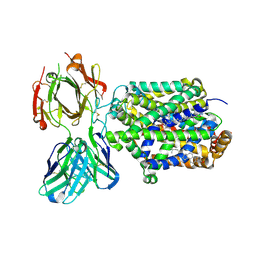 | | X-ray structure of Drosophila dopamine transporter with subsiteB mutations (D121G/S426M) bound to cocaine | | Descriptor: | Antibody fragment heavy chain-protein, 9D5-heavy chain, Antibody fragment light chain-protein, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
4FOO
 
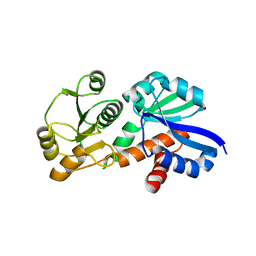 | |
1HK7
 
 | | Middle Domain of HSP90 | | Descriptor: | CADMIUM ION, HEAT SHOCK PROTEIN HSP82, MAGNESIUM ION | | Authors: | Meyer, P, Prodromou, C, Roe, S.M, Pearl, L.H. | | Deposit date: | 2003-03-06 | | Release date: | 2004-01-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Analysis of the Middle Segment of Hsp90. Implications for ATP Hydrolysis and Client Protein and Cochaperone Interactions
Mol.Cell, 11, 2003
|
|
4XPT
 
 | | X-ray structure of Drosophila dopamine transporter with subsiteB mutations D121G/S426M and EL2 deletion of 162-201 in complex with substrate analogue 3,4 dichlorophen ethylamine | | Descriptor: | 2-(3,4-dichlorophenyl)ethanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-17 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
4FOU
 
 | | Structure of the PilZ-FimX(EAL domain)-c-di-GMP complex responsible for the regulation of bacterial Type IV pilus biogenesis | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, FimX, ... | | Authors: | Farah, C.S, Guzzo, C.R. | | Deposit date: | 2012-06-21 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the PilZ-FimXEAL-c-di-GMP Complex Responsible for the Regulation of Bacterial Type IV Pilus Biogenesis.
J.Mol.Biol., 425, 2013
|
|
1HKO
 
 | | NMR structure of bovine cytochrome b5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Muskett, F.W, Whitford, D. | | Deposit date: | 2003-03-10 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of Bovine Ferricytochrome B5 Determined Using Heteronuclear NMR Methods.
J.Mol.Biol., 258, 1996
|
|
4FGE
 
 | | Structure of the effector protein Tse1 from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, GLYCEROL, THIOCYANATE ION, ... | | Authors: | Benz, J, Sendlmeier, C, Barends, T.R.M, Meinhart, A. | | Deposit date: | 2012-06-04 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights into the Effector - Immunity System Tse1/Tsi1 from Pseudomonas aeruginosa.
Plos One, 7, 2012
|
|
4FR5
 
 | | Crystal Structure of Shikimate Dehydrogenase (aroE) Y210S Mutant from Helicobacter pylori in Complex with Shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, Shikimate dehydrogenase | | Authors: | Cheng, W.C, Chen, T.J, Wang, W.C. | | Deposit date: | 2012-06-26 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: |
|
|
5EXW
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 7-deaza-ATP | | Descriptor: | 7-deazaadenosine-5'-triphosphate, 78 kDa glucose-regulated protein | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-24 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
8F6Q
 
 | | CryoEM structure of designed modular protein oligomer C8-71 | | Descriptor: | C8-71 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2022-11-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Modulation of FGF pathway signaling and vascular differentiation using designed oligomeric assemblies.
Biorxiv, 2023
|
|
8F39
 
 | | Yeast ATP synthase in conformation-2, at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-09 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8F29
 
 | | Yeast ATP synthase in conformation-1 at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-07 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5F1X
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with ATP | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-TRIPHOSPHATE | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
2ZDY
 
 | | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kawamoto, M, Shiromizu, I, Kukimoto-Niino, M, Tokmakov, A, Terada, T, Shirouzu, M, Matsusue, T, Yokoyama, S. | | Deposit date: | 2007-12-01 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3D38
 
 | | Crystal structure of new trigonal form of photosynthetic reaction center from Blastochloris viridis. Crystals grown in microfluidics by detergent capture. | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Li, L, Nachtergaele, S.H.M, Seddon, A.M, Tereshko, V, Ponomarenko, N, Ismagilov, R.F, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Simple host-guest chemistry to modulate the process of concentration and crystallization of membrane proteins by detergent capture in a microfluidic device.
J.Am.Chem.Soc., 130, 2008
|
|
5EY4
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 2'-deoxy-ATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 78 kDa glucose-regulated protein | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-24 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
8FEF
 
 | | Structure of Mce1 transporter from Mycobacterium smegmatis (Map0) | | Descriptor: | ABC transporter, ATP-binding protein,Green fluorescent protein chimera, ABC-transporter integral membrane protein, ... | | Authors: | Chen, J, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structure of an endogenous mycobacterial MCE lipid transporter.
Nature, 620, 2023
|
|
8FED
 
 | | Structure of Mce1-LucB complex from Mycobacterium smegmatis (Map1) | | Descriptor: | ABC transporter, ATP-binding protein,Green fluorescent protein chimera, ABC-transporter integral membrane protein, ... | | Authors: | Chen, J, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structure of an endogenous mycobacterial MCE lipid transporter.
Nature, 620, 2023
|
|
8FEE
 
 | | Structure of Mce1 transporter from Mycobacterium smegmatis in the absence of LucB (Map2) | | Descriptor: | ABC transporter, ATP-binding protein,Green fluorescent protein chimera, ABC-transporter integral membrane protein, ... | | Authors: | Chen, J, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of an endogenous mycobacterial MCE lipid transporter.
Nature, 620, 2023
|
|
5F0X
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 2'-deoxy-ADP and inorganic phosphate | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, 78 kDa glucose-regulated protein, ... | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-28 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
2IQX
 
 | | Rat Phosphatidylethanolamine-Binding Protein Containing the S153E Mutation in the Complex with o-Phosphorylethanolamine | | Descriptor: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Phosphatidylethanolamine-binding protein 1 | | Authors: | Kim, Y, Joachimiak, G, Clark, M.C, Rosner, M, Joachimiak, A. | | Deposit date: | 2006-10-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rat Phosphatidylethanolamine-Binding Crystal structure of Protein Containing the S153E Mutation in the Complex with o-Phosphorylethanolamine
To be Published
|
|
5EX5
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 7-deaza-ADP and inorganic phosphate | | Descriptor: | 7-deazaadenosine-5'-diphosphate, 78 kDa glucose-regulated protein, MAGNESIUM ION, ... | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
