5IIS
 
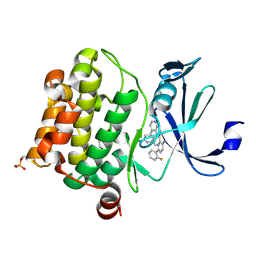 | | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl-amide scaffold | | Descriptor: | 3-amino-N-(2'-amino-6'-methyl[4,4'-bipyridin]-3-yl)-6-(2-fluorophenyl)pyridine-2-carboxamide, DI(HYDROXYETHYL)ETHER, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C, Bussiere, D, Burger, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl carboxamide scaffold.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6K4I
 
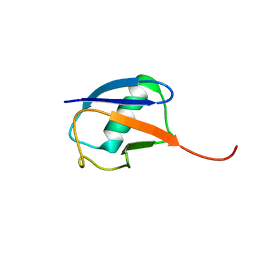 | | The partially disordered conformation of ubiquitin (Q41N variant) | | Descriptor: | ubiquitin | | Authors: | Wakamoto, T, Ikeya, T, Kitazawa, S, Baxter, N.J, Williamson, M.P, Kitahara, R. | | Deposit date: | 2019-05-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Paramagnetic relaxation enhancement-assisted structural characterization of a partially disordered conformation of ubiquitin.
Protein Sci., 28, 2019
|
|
1KWD
 
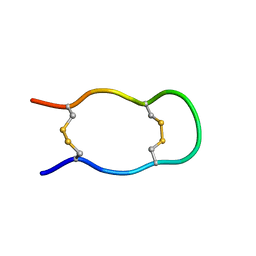 | | SOLUTION STRUCTURE OF THE CENTRAL CONSERVED REGION OF HUMAN RESPIRATORY SYNCYTIAL VIRUS ATTACHMENT GLYCOPROTEIN G 187 | | Descriptor: | MAJOR SURFACE GLYCOPROTEIN G | | Authors: | Sugawara, M, Czaplicki, J, Ferrage, J, Haeuw, J.F, Power, U.F, Corvaia, N, Nguyen, T, Beck, A, Milon, A. | | Deposit date: | 2002-01-29 | | Release date: | 2003-06-17 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structure-antigenicity relationship studies of the central conserved region of human respiratory syncytial virus protein G.
J.Pept.Res., 60, 2002
|
|
1B0E
 
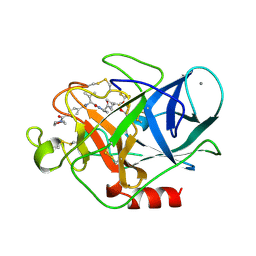 | | CRYSTAL STRUCTURE OF PORCINE PANCREATIC ELASTASE WITH MDL 101,146 | | Descriptor: | 1-{3-METHYL-2-[4-(MORPHOLINE-4-CARBONYL)-BENZOYLAMINO]-BUTYRYL}-PYRROLIDINE-2-CARBOXYLIC ACID (3,3,4,4,4-PENTAFLUORO-1-ISOPROPYL-2-OXO-BUTYL)-AMIDE, CALCIUM ION, PROTEIN (ELASTASE) | | Authors: | Schreuder, H.A, Metz, W.A, Peet, N.P, Pelton, J.T, Tardif, C. | | Deposit date: | 1998-11-09 | | Release date: | 1998-11-18 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of human neutrophil elastase. 4. Design, synthesis, X-ray crystallographic analysis, and structure-activity relationships for a series of P2-modified, orally active peptidyl pentafluoroethyl ketones.
J.Med.Chem., 41, 1998
|
|
1KWE
 
 | | SOLUTION STRUCTURE OF THE CENTRAL CONSERVED REGION OF HUMAN RESPIRATORY SYNCYTIAL VIRUS ATTACHMENT GLYCOPROTEIN G | | Descriptor: | MAJOR SURFACE GLYCOPROTEIN G | | Authors: | Sugawara, M, Czaplicki, J, Ferrage, J, Haeuw, J.F, Power, U.F, Corvaia, N, Nguyen, T, Beck, A, Milon, A. | | Deposit date: | 2002-01-29 | | Release date: | 2003-06-17 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structure-antigenicity relationship studies of the central conserved region of human respiratory syncytial virus protein G.
J.Pept.Res., 60, 2002
|
|
1DI7
 
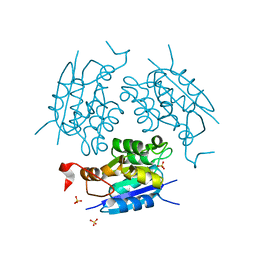 | | 1.60 ANGSTROM CRYSTAL STRUCTURE OF THE MOLYBDENUM COFACTOR BIOSYNTHESIS PROTEIN MOGA FROM ESCHERICHIA COLI | | Descriptor: | MOLYBDENUM COFACTOR BIOSYNTHETIC ENZYME, SULFATE ION | | Authors: | Liu, M.T.W, Wuebbens, M.M, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 1999-11-29 | | Release date: | 2000-01-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the gephyrin-related molybdenum cofactor biosynthesis protein MogA from Escherichia coli.
J.Biol.Chem., 275, 2000
|
|
1DI6
 
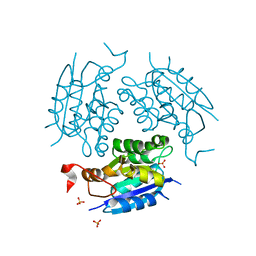 | | 1.45 A CRYSTAL STRUCTURE OF THE MOLYBDENUMM COFACTOR BIOSYNTHESIS PROTEIN MOGA FROM ESCHERICHIA COLI | | Descriptor: | MOLYBDENUM COFACTOR BIOSYNTHETIC ENZYME, SULFATE ION | | Authors: | Liu, M.T.W, Wuebbens, M.M, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 1999-11-29 | | Release date: | 2000-01-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the gephyrin-related molybdenum cofactor biosynthesis protein MogA from Escherichia coli.
J.Biol.Chem., 275, 2000
|
|
5VJ4
 
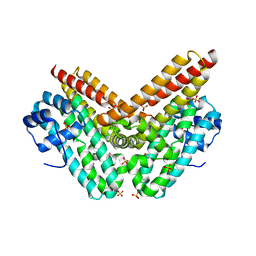 | | Crystal structure of a putative surface protein encoded by BTA121 | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Luo, Z, Asojo, O. | | Deposit date: | 2017-04-18 | | Release date: | 2017-11-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Borrelia turicatae protein, BTA121, a differentially regulated gene in the tick-mammalian transmission cycle of relapsing fever spirochetes.
Sci Rep, 7, 2017
|
|
3TTB
 
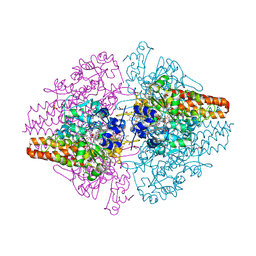 | | Structure of the Thioalkalivibrio paradoxus cytochrome c nitrite reductase in complex with sulfite | | Descriptor: | CALCIUM ION, COBALT (II) ION, Eight-heme nitrite reductase, ... | | Authors: | Polyakov, K.M, Trofimov, A.A, Tikhonova, T.V, Tikhonov, A.V, Dorovatovskii, P.V, Popov, V.O. | | Deposit date: | 2011-09-14 | | Release date: | 2011-10-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparative structural and functional analysis of two octaheme nitrite reductases from closely related Thioalkalivibrio species.
Febs J., 279, 2012
|
|
7LVI
 
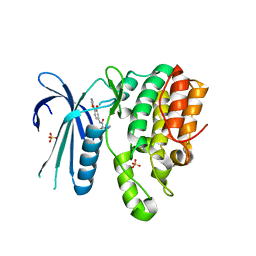 | | CRYSTAL STRUCTURE OF AP2 ASSOCIATED KINASE 1 ISOFORM 1 COMPLEXED WITH LIGAND (2R)-2-AMINO-N-[3-METHOXY-4- (1,3-OXAZOL-5-YL)PHENYL]-4-METHYLPENTANAMIDE | | Descriptor: | (2R,3R)-N-[3-methoxy-4-(1,3-oxazol-5-yl)phenyl]-3-(propan-2-yl)piperidine-2-carboxamide, 5-[(4-aminopiperidin-1-yl)methyl]-N-{3-[5-(propan-2-yl)-1,3,4-thiadiazol-2-yl]phenyl}pyrrolo[2,1-f][1,2,4]triazin-4-amine, AP2-associated protein kinase 1, ... | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery, Structure-Activity Relationships, and In Vivo Evaluation of Novel Aryl Amides as Brain Penetrant Adaptor Protein 2-Associated Kinase 1 (AAK1) Inhibitors for the Treatment of Neuropathic Pain.
J.Med.Chem., 64, 2021
|
|
7LVH
 
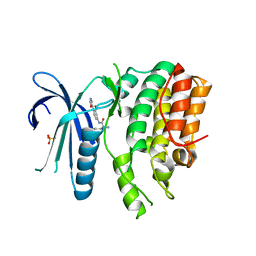 | | CRYSTAL STRUCTURE OF AP2 ASSOCIATED KINASE 1 ISOFORM 1 COMPLEXED WITH LIGAND N-[3-METHOXY-4-(1,3-OXAZOL-5-YL)PHENYL]-3-(PROPAN-2-YL)PIPERIDINE-2-CARBOXAMIDE | | Descriptor: | 5-[(4-aminopiperidin-1-yl)methyl]-N-{3-[5-(propan-2-yl)-1,3,4-thiadiazol-2-yl]phenyl}pyrrolo[2,1-f][1,2,4]triazin-4-amine, AP2-associated protein kinase 1, N-[3-methoxy-4-(1,3-oxazol-5-yl)phenyl]-D-leucinamide, ... | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery, Structure-Activity Relationships, and In Vivo Evaluation of Novel Aryl Amides as Brain Penetrant Adaptor Protein 2-Associated Kinase 1 (AAK1) Inhibitors for the Treatment of Neuropathic Pain.
J.Med.Chem., 64, 2021
|
|
4ELE
 
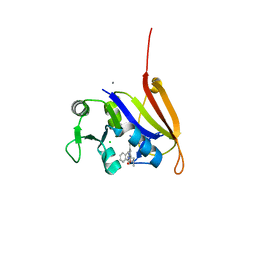 | | Structure-activity relationship guides enantiomeric preference among potent inhibitors of B. anthracis dihydrofolate reductase | | Descriptor: | (2E)-3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1S)-1-(propan-2-yl)phthalazin-2(1H)-yl]prop-2-en -1-one, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bourne, C.R, Barrow, W.W. | | Deposit date: | 2012-04-10 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-activity relationship for enantiomers of potent inhibitors of B. anthracis dihydrofolate reductase.
Biochim.Biophys.Acta, 1834, 2013
|
|
4ELG
 
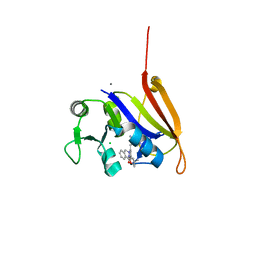 | | Structure-activity relationship guides enantiomeric preference among potent inhibitors of B. anthracis dihydrofolate reductase | | Descriptor: | (2E)-3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1R)-1-(2-methylpropyl)phthalazin-2(1H)-yl]prop-2 -en-1-one, (2E)-3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1S)-1-(2-methylpropyl)phthalazin-2(1H)-yl]prop-2-en-1-one, CALCIUM ION, ... | | Authors: | Bourne, C.R, Barrow, W.W. | | Deposit date: | 2012-04-10 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure-activity relationship for enantiomers of potent inhibitors of B. anthracis dihydrofolate reductase.
Biochim.Biophys.Acta, 1834, 2013
|
|
5BOJ
 
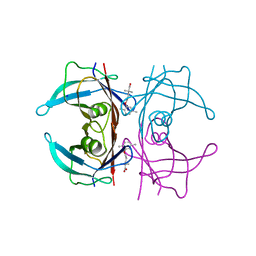 | | Crystal Structure of Human Transthyretin in Complex with Gemfibrozil | | Descriptor: | 5-(2,5-dimethylphenoxy)-2,2-dimethylpentanoic acid, SODIUM ION, Transthyretin | | Authors: | Begum, A, Olofsson, A, Sauer-Eriksson, A.E. | | Deposit date: | 2015-05-27 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Enthalpic Forces Correlate with the Selectivity of Transthyretin-Stabilizing Ligands in Human Plasma.
J.Med.Chem., 58, 2015
|
|
1IYC
 
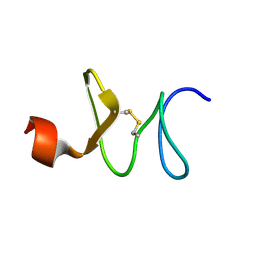 | | Solution structure of antifungal peptide, scarabaecin | | Descriptor: | scarabaecin | | Authors: | Hemmi, H, Ishibashi, J, Tomie, T, Yamakawa, M. | | Deposit date: | 2002-08-05 | | Release date: | 2003-06-24 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for New Pattern of Conserved Amino Acid Residues Related to Chitin-binding in the Antifungal Peptide from the Coconut Rhinoceros Beetle Oryctes rhinoceros
J.BIOL.CHEM., 278, 2003
|
|
2L92
 
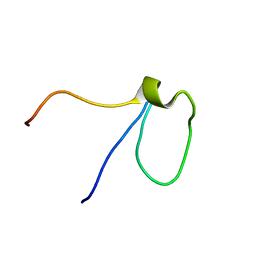 | |
7MS7
 
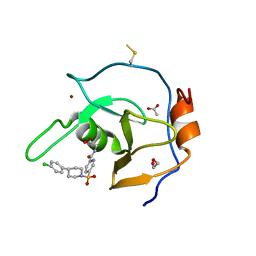 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (5-((4-(4-chlorophenyl)piperidin-1-yl)sulfonyl)picolinoyl)glycine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, N-{5-[4-(4-chlorophenyl)piperidine-1-sulfonyl]pyridine-2-carbonyl}glycine, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
4GCE
 
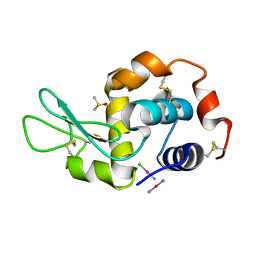 | |
3HHX
 
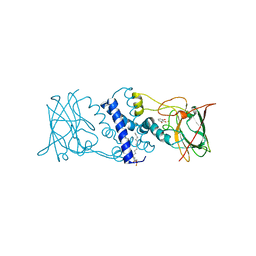 | | Crystal structure determination of Catechol 1,2-Dioxygenase from Rhodococcus opacus 1CP in complex with pyrogallol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BENZENE-1,2,3-TRIOL, Catechol 1,2-dioxygenase, ... | | Authors: | Matera, I, Ferraroni, M, Briganti, F. | | Deposit date: | 2009-05-18 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
7MS5
 
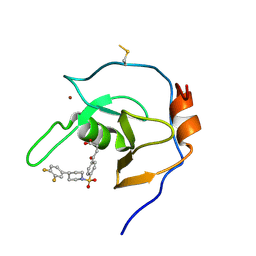 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 4-(4-(4-(3,4-difluoro-phenyl)-piperidin-1-ylsulfonyl)-phenyl)-4-oxo-butanoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[4-(3,4-difluorophenyl)piperidine-1-sulfonyl]phenyl}-4-oxobutanoic acid, CALCIUM ION, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7MS6
 
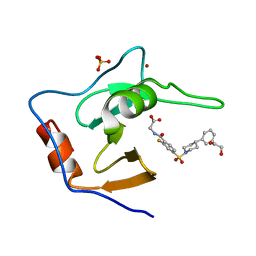 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (2-fluoro-4-((4-phenylpiperidin-1-yl)sulfonyl)benzoyl)glycine | | Descriptor: | 1,2-ETHANEDIOL, N-[2-fluoro-4-(4-phenylpiperidine-1-sulfonyl)benzoyl]glycine, SULFATE ION, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
3HHY
 
 | | Crystal structure determination of Catechol 1,2-Dioxygenase from Rhodococcus opacus 1CP in complex with catechol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, CATECHOL, CHLORIDE ION, ... | | Authors: | Matera, I, Ferraroni, M, Kolomytseva, M, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-05-18 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
3HGI
 
 | | Crystal structure of Catechol 1,2-Dioxygenase from the gram-positive Rhodococcus opacus 1CP | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BENZOIC ACID, CARBONATE ION, ... | | Authors: | Ferraroni, M, Matera, I, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-05-14 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
4GCB
 
 | |
4GCF
 
 | |
