3ITJ
 
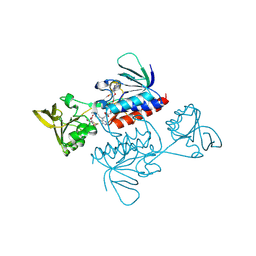 | | Crystal structure of Saccharomyces cerevisiae thioredoxin reductase 1 (Trr1) | | Descriptor: | CITRIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase 1 | | Authors: | Oliveira, M.A, Discola, K.F, Alves, S.V, Medrano, F.J, Guimaraes, B.G, Netto, L.E.S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-03-31 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the specificity of thioredoxin reductase-thioredoxin interactions. A structural and functional investigation of the yeast thioredoxin system.
Biochemistry, 49, 2010
|
|
2GD1
 
 | |
3RN1
 
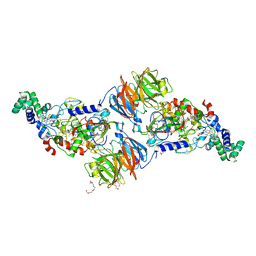 | |
3WIC
 
 | | Structure of a substrate/cofactor-unbound glucose dehydrogenase | | Descriptor: | Glucose 1-dehydrogenase, PENTAETHYLENE GLYCOL, S-1,2-PROPANEDIOL, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1DOH
 
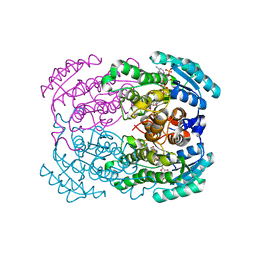 | | STRUCTURE OF TRIHYDROXYNAPHTHALENE REDUCTASE IN COMPLEX WITH NADPH AND 4-NITRO-INDEN-1-ONE | | Descriptor: | 4-NITRO-INDEN-1-ONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIHYDROXYNAPHTHALENE REDUCTASE | | Authors: | Liao, D.I, Basarab, G.S, Gatenby, A.A, Jordan, D.B. | | Deposit date: | 1999-12-20 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of trihydroxynaphthalene reductase-fungicide complexes: implications for structure-based design and catalysis.
Structure, 9, 2001
|
|
1FW1
 
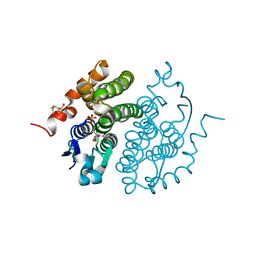 | | Glutathione transferase zeta/maleylacetoacetate isomerase | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLUTATHIONE, GLUTATHIONE TRANSFERASE ZETA, ... | | Authors: | Polekhina, G, Board, P.G, Blackburn, A.C, Parker, M.W. | | Deposit date: | 2000-09-20 | | Release date: | 2001-09-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of maleylacetoacetate isomerase/glutathione transferase zeta reveals the molecular basis for its remarkable catalytic promiscuity.
Biochemistry, 40, 2001
|
|
1A7A
 
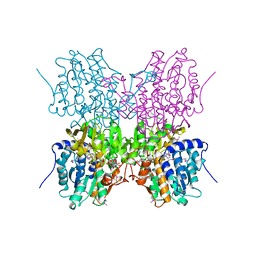 | | STRUCTURE OF HUMAN PLACENTAL S-ADENOSYLHOMOCYSTEINE HYDROLASE: DETERMINATION OF A 30 SELENIUM ATOM SUBSTRUCTURE FROM DATA AT A SINGLE WAVELENGTH | | Descriptor: | (1'R,2'S)-9-(2-HYDROXY-3'-KETO-CYCLOPENTEN-1-YL)ADENINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-ADENOSYLHOMOCYSTEINE HYDROLASE | | Authors: | Turner, M.A, Yuan, C.-S, Borchardt, R.T, Hershfield, M.S, Smith, G.D, Howell, P.L. | | Deposit date: | 1998-03-10 | | Release date: | 1999-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure determination of selenomethionyl S-adenosylhomocysteine hydrolase using data at a single wavelength.
Nat.Struct.Biol., 5, 1998
|
|
1M1N
 
 | | Nitrogenase MoFe protein from Azotobacter vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(7)-MO-S(9)-N CLUSTER, ... | | Authors: | Einsle, O, Tezcan, F.A, Andrade, S.L.A, Schmid, B, Yoshida, M, Howard, J.B, Rees, D.C. | | Deposit date: | 2002-06-19 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Nitrogenase MoFe-protein at 1.16 A resolution: a central ligand in the FeMo-cofactor.
Science, 297, 2002
|
|
4S3A
 
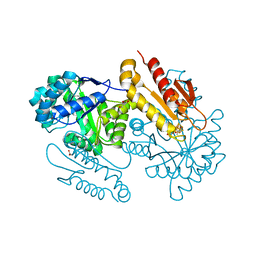 | | IspG in complex with Intermediate I | | Descriptor: | (3S)-1,3-dihydroxy-4-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}-2-methylbut-2-ylium, carbokation intermediate, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
6MO4
 
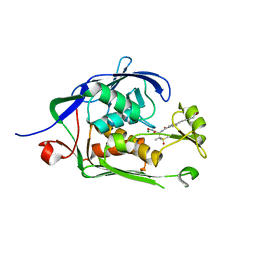 | | Co-Crystal structure of P. aeruginosa LpxC-50067 complex | | Descriptor: | MAGNESIUM ION, N-[(2R)-1-(hydroxyamino)-3-methyl-3-(methylsulfonyl)-1-oxobutan-2-yl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase | | Authors: | Stein, A.J, Assar, Z, Holt, M.C, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
4JMV
 
 | |
1GEG
 
 | | CRYATAL STRUCTURE ANALYSIS OF MESO-2,3-BUTANEDIOL DEHYDROGENASE | | Descriptor: | ACETOIN REDUCTASE, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | Otagiri, M, Kurisu, G, Ui, S, Kusunoki, M. | | Deposit date: | 2000-11-10 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of meso-2,3-butanediol dehydrogenase in a complex with NAD+ and inhibitor mercaptoethanol at 1.7 A resolution for understanding of chiral substrate recognition mechanisms.
J.Biochem., 129, 2001
|
|
1QLU
 
 | |
5W7D
 
 | | Murine acyloxyacyl hydrolase (AOAH), S262A mutant | | Descriptor: | 1,2-DISTEAROYL-SN-GLYCERO-3-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2017-06-19 | | Release date: | 2018-01-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the mammalian lipopolysaccharide detoxifier.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2WWD
 
 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae in complex with pneummococcal peptidoglycan fragment | | Descriptor: | 1,4-BETA-N-ACETYLMURAMIDASE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, ALANINE, ... | | Authors: | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L. | | Deposit date: | 2009-10-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3ORV
 
 | |
4OTJ
 
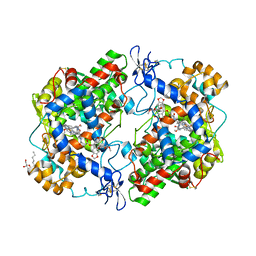 | | The complex of murine cyclooxygenase-2 with a conjugate of indomefathin and podophyllotoxin, N-{(succinylpodophyllotoxinyl)but-4-yl}-2-{1-(4-chlorobenzoyl)-5-methoxy-2-methyl-1H-indol-3-yl}acetamide | | Descriptor: | (5S,5aS,8aS,9S)-8-oxo-9-(3,4,5-trimethoxyphenyl)-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol-5-yl 4-{[4-({[1-(4-chlorobenzoyl)-5-methoxy-2-methyl-1H-indol-3-yl]acetyl}amino)butyl]amino}-4-oxobutanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, S, Uddin, M.J, Banerjee, S, Marnett, L.J. | | Deposit date: | 2014-02-13 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Antitumor Activity of Cytotoxic Cyclooxygenase-2 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
3FMX
 
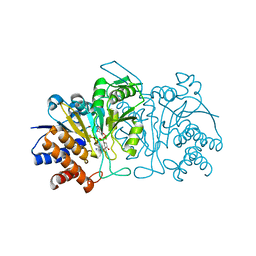 | |
2WHP
 
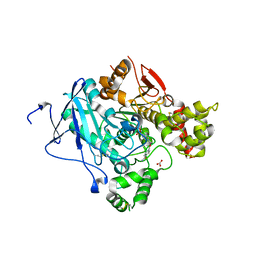 | | Crystal structure of acetylcholinesterase, phosphonylated by sarin and in complex with HI-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Ekstrom, F, Hornberg, A, Artursson, E, Hammarstrom, L.G, Schneider, G, Pang, Y.P. | | Deposit date: | 2009-05-06 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Hi-6Sarin-Acetylcholinesterase Determined by X-Ray Crystallography and Molecular Dynamics Simulation: Reactivator Mechanism and Design.
Plos One, 4, 2009
|
|
3CCT
 
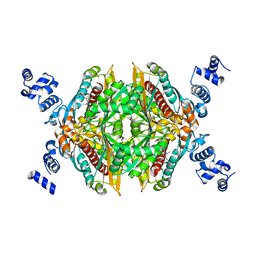 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-[2-(4-fluorophenyl)-4-[(2-hydroxyphenyl)carbamoyl]-5-(1-methylethyl)-3-phenyl-1H-pyrrol-1-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
1EL5
 
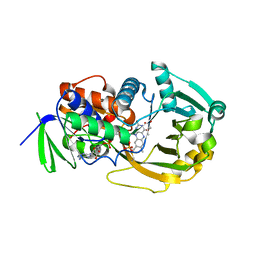 | | COMPLEX OF MONOMERIC SARCOSINE OXIDASE WITH THE INHIBITOR DIMETHYLGLYCINE | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, N,N-DIMETHYLGLYCINE, ... | | Authors: | Wagner, M.A, Trickey, P, Chen, Z.-W, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2000-03-13 | | Release date: | 2000-12-06 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Monomeric sarcosine oxidase: 1. Flavin reactivity and active site binding determinants.
Biochemistry, 39, 2000
|
|
3CD5
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-[3-(biphenyl-4-ylcarbamoyl)-2-ethyl-5,6,7,8-tetrahydrocyclohepta[b]pyrrol-1(4H)-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
3G91
 
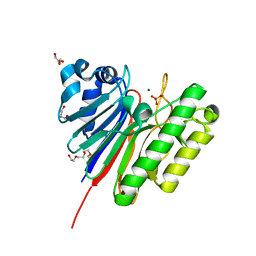 | | 1.2 Angstrom structure of the exonuclease III homologue Mth0212 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Exodeoxyribonuclease, GLYCEROL, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-12 | | Release date: | 2010-03-09 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA.
J.Mol.Biol., 399, 2010
|
|
4S23
 
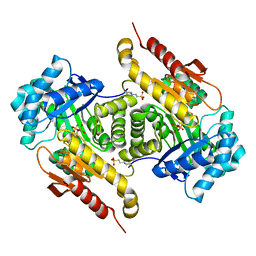 | | Structure of the GcpE-HMBPP complex from Thermus thermophilius | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, ... | | Authors: | Rekittke, I, Warkentin, E, Jomaa, H, Ermler, E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the GcpE-HMBPP complex from Thermus thermophilius.
Biochem.Biophys.Res.Commun., 458, 2015
|
|
4S39
 
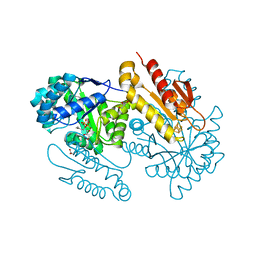 | | IspG in complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, GLYCEROL, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Guodong, R, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
