8VAP
 
 | |
8VAM
 
 | |
8VAS
 
 | |
5F15
 
 | | Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose (L-Ara4N) transferase, CHLORIDE ION, ... | | Authors: | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
6VU7
 
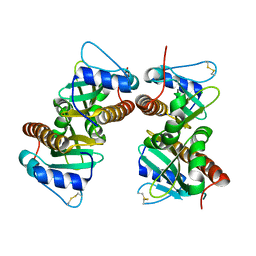 | | Crystal structure of YbjN, a putative transcription regulator from E. coli | | Descriptor: | CHLORIDE ION, YbjN protein | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of YbjN, a putative transcription regulator from E. coli
To Be Published
|
|
7SQD
 
 | |
6G2T
 
 | | human cardiac myosin binding protein C C1 Ig-domain bound to native cardiac thin filament | | Descriptor: | Actin, cytoplasmic 2, Myosin-binding protein C, ... | | Authors: | Risi, C, Belknap, B, Forgacs, E, Harris, S.P, Schroder, G.F, White, H.D, Galkin, V.E. | | Deposit date: | 2018-03-23 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | N-Terminal Domains of Cardiac Myosin Binding Protein C Cooperatively Activate the Thin Filament.
Structure, 26, 2018
|
|
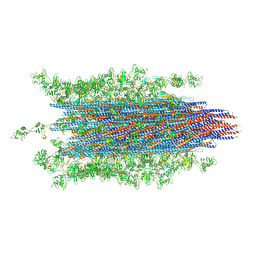
6MRU
 
 | | 13-meric ClyA pore complex | | Descriptor: | Hemolysin E, chromosomal | | Authors: | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
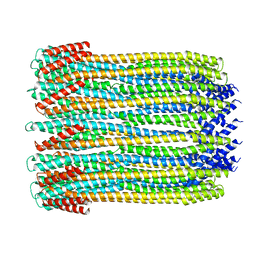
4QR8
 
 | | Crystal Structure of E coli pepQ | | Descriptor: | MAGNESIUM ION, Xaa-Pro dipeptidase | | Authors: | Pingwei, L. | | Deposit date: | 2014-06-30 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural basis of substrate selectivity of E. coli prolidase.
Plos One, 9, 2014
|
|
4RFO
 
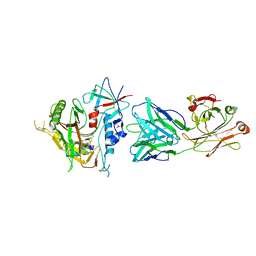 | | Crystal structure of the ADCC-Potent Antibody N60-I3 Fab in complex with HIV-1 Clade A/E gp120 and M48u1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade A/E gp120, N60-i3 Fab heavy chain, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2014-09-26 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
6MWR
 
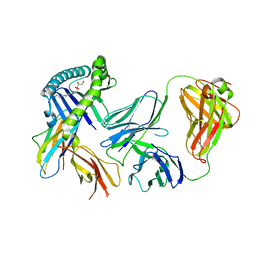 | | Recognition of MHC-like molecule | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Le Nours, J, Rossjohn, J. | | Deposit date: | 2018-10-30 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A class of gamma delta T cell receptors recognize the underside of the antigen-presenting molecule MR1.
Science, 366, 2019
|
|
8A1V
 
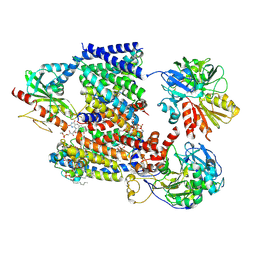 | | Sodium pumping NADH-quinone oxidoreductase with substrate Q2 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1W
 
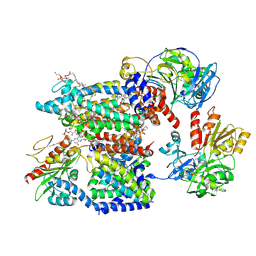 | | Sodium pumping NADH-quinone oxidoreductase with substrate Q1 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
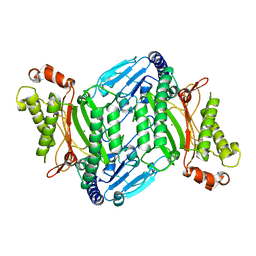
4R9M
 
 | | Crystal structure of spermidine N-acetyltransferase from Escherichia coli | | Descriptor: | MAGNESIUM ION, Spermidine N(1)-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Grimshaw, S, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Analysis of crystalline and solution states of ligand-free spermidine N-acetyltransferase (SpeG) from Escherichia coli.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8A1T
 
 | | Sodium pumping NADH-quinone oxidoreductase | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1X
 
 | | Sodium pumping NADH-quinone oxidoreductase with inhibitor DQA | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1Y
 
 | | Sodium pumping NADH-quinone oxidoreductase with inhibitor HQNO | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8PMZ
 
 | | HEV gt3 P domain in complex with glycan-insensitive nAb p61.15 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pro-secreted protein ORF2, scFv_p61.15 | | Authors: | Ssebyatika, G, Krey, T. | | Deposit date: | 2023-06-29 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | A novel class of broadly neutralizing hepatitis E virus-specific human antibodies
To Be Published
|
|
8PMY
 
 | |
4RFN
 
 | | Crystal structure of ADCC-potent Rhesus macaque ANTIBODY JR4 in complex with HIV-1 CLADE A/E GP120 and M48 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB HEAVY CHAIN OF ADCC ANTI-HIV-1 ANTIBODY JR4, FAB LIGHT CHAIN OF ADCC ANTI-HIV-1 ANTIBODY JR4, ... | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2014-09-26 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
3D74
 
 | | Crystal structure of a pheromone binding protein mutant D35A, from Apis mellifera, soaked at pH 5.5 | | Descriptor: | N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
8A1U
 
 | | Sodium pumping NADH-quinone oxidoreductase with substrates NADH and Q2 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6NPS
 
 | | Crystal structure of GH115 enzyme AxyAgu115A from Amphibacillus xylanus | | Descriptor: | AxyAgu115A, CHLORIDE ION, GLYCEROL | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Yan, R, Master, E, Savchenko, A. | | Deposit date: | 2019-01-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural characterization of the family GH115 alpha-glucuronidase from Amphibacillus xylanus yields insight into its coordinated action with alpha-arabinofuranosidases.
N Biotechnol, 2021
|
|
3CT7
 
 | | Crystal structure of D-allulose 6-phosphate 3-epimerase from Escherichia Coli K-12 | | Descriptor: | D-allulose-6-phosphate 3-epimerase, MAGNESIUM ION, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-04-11 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for substrate specificity in phosphate binding (beta/alpha)8-barrels: D-allulose 6-phosphate 3-epimerase from Escherichia coli K-12.
Biochemistry, 47, 2008
|
|
8B1V
 
 | | Dihydroprecondylocarpine acetate synthase 2 from Tabernanthe iboga | | Descriptor: | Dihydroprecondylocarpine acetate synthase 2, ZINC ION, precondylocarpine acetate | | Authors: | Langley, C, Basquin, J, Caputi, L, O'Connor, S.E. | | Deposit date: | 2022-09-12 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Expansion of the Catalytic Repertoire of Alcohol Dehydrogenases in Plant Metabolism.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
