6MGI
 
 | |
2RPR
 
 | | Solution structure of the fifth FLYWCH domain of FLYWCH-type zinc finger-containing protein 1 | | Descriptor: | FLYWCH-type zinc finger-containing protein 1, ZINC ION | | Authors: | Enomoto, M, Saito, K, Tochio, N, Kigawa, T, Yokoyama, S, Nameki, N. | | Deposit date: | 2008-07-24 | | Release date: | 2009-08-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fifth FLYWCH domain of FLYWCH-type zinc finger-containing protein 1
To be Published
|
|
5VW1
 
 | | Crystal structure of SpyCas9-sgRNA-AcrIIA4 ternary complex | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, H, Patel, D.J. | | Deposit date: | 2017-05-21 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Inhibition Mechanism of an Anti-CRISPR Suppressor AcrIIA4 Targeting SpyCas9.
Mol. Cell, 67, 2017
|
|
4S24
 
 | | 1.7 Angstrom Crystal Structure of of Putative Modulator of Drug Activity (apo- form) from Yersinia pestis CO92 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Modulator of drug activity B, PENTAETHYLENE GLYCOL, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-01-19 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Crystal Structure of of Putative Modulator of Drug Activity (apo- form) from Yersinia pestis CO92.
TO BE PUBLISHED
|
|
1EGL
 
 | |
3BO9
 
 | |
4YH3
 
 | | Crystal structure of human BRD4(1) in complex with 4-[(2E)-3-(4-methoxyphenyl)-2-phenylprop-2-enoyl]-3,4-dihydroquinoxalin-2(1H)-one (compound 19a) | | Descriptor: | 4-[(2E)-3-(4-methoxyphenyl)-2-phenylprop-2-enoyl]-3,4-dihydroquinoxalin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | White, A, Lakshminarasimhan, D, Suto, R.K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of a new chemical series of BRD4(1) inhibitors using protein-ligand docking and structure-guided design.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6JRK
 
 | | The structure of co-crystals of 8r-B-EGFR WT complex | | Descriptor: | 6-(2-chloranyl-3-fluoranyl-phenyl)-5-methyl-2-[[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino]-8-[(3S)-1-propanoylpiperidin-3-yl]pyrido[2,3-d]pyrimidin-7-one, Epidermal growth factor receptor | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2019-04-04 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structure-Based Design of 5-Methylpyrimidopyridone Derivatives as New Wild-Type Sparing Inhibitors of the Epidermal Growth Factor Receptor Triple Mutant (EGFRL858R/T790M/C797S).
J.Med.Chem., 62, 2019
|
|
4LQT
 
 | | 1.10A resolution crystal structure of a superfolder green fluorescent protein (W57A) mutant | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein | | Authors: | Lovell, S, Xia, Y, Vo, B, Battaile, K.P, Egan, C, Karanicolas, J. | | Deposit date: | 2013-07-19 | | Release date: | 2013-12-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The designability of protein switches by chemical rescue of structure: mechanisms of inactivation and reactivation.
J.Am.Chem.Soc., 135, 2013
|
|
6JRJ
 
 | | The structure of co-crystals of 8r-B-EGFR T790M/C797S complex | | Descriptor: | 6-(2-chloranyl-3-fluoranyl-phenyl)-5-methyl-2-[[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino]-8-[(3S)-1-propanoylpiperidin-3-yl]pyrido[2,3-d]pyrimidin-7-one, Epidermal growth factor receptor | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2019-04-04 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.943 Å) | | Cite: | Structure-Based Design of 5-Methylpyrimidopyridone Derivatives as New Wild-Type Sparing Inhibitors of the Epidermal Growth Factor Receptor Triple Mutant (EGFRL858R/T790M/C797S).
J.Med.Chem., 62, 2019
|
|
5GGG
 
 | |
5GGN
 
 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GlcNAc-beta-pNP | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5H0N
 
 | |
5CIB
 
 | | Complex of yeast cytochrome c peroxidase (W191G) bound to 2,4-dimethylaniline with iso-1 cytochrome c | | Descriptor: | 2,4-dimethylaniline, Cytochrome c iso-1, Cytochrome c peroxidase, ... | | Authors: | Crane, B.R, Payne, T.M. | | Deposit date: | 2015-07-11 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Constraints on the Radical Cation Center of Cytochrome c Peroxidase for Electron Transfer from Cytochrome c.
Biochemistry, 55, 2016
|
|
2GZ4
 
 | | 1.5 A Crystal Structure of a Protein of Unknown Function ATU1052 from Agrobacterium tumefaciens | | Descriptor: | Hypothetical protein Atu1052 | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-05-10 | | Release date: | 2006-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein Atu1052 from Agrobacterium tumefaciens
To be Published
|
|
1SH8
 
 | | 1.5 A Crystal Structure of a Protein of Unknown Function PA5026 from Pseudomonas aeruginosa, Probable Thioesterase | | Descriptor: | hypothetical protein PA5026 | | Authors: | Zhang, R, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein PA5026 from Pseudomonas aeruginosa
To be Published
|
|
4LSM
 
 | |
4LYI
 
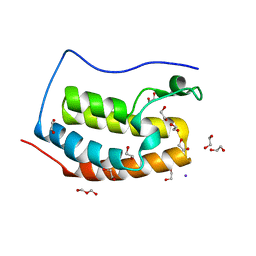 | | Crystal Structure of apo-BRD4(1) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, IODIDE ION, ... | | Authors: | Wohlwend, D. | | Deposit date: | 2013-07-31 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 4-Acyl pyrroles: mimicking acetylated lysines in histone code reading.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
6KEV
 
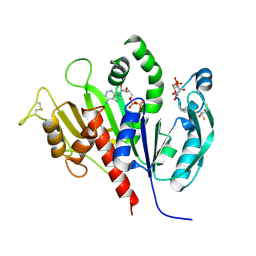 | | Reduced phosphoribulokinase from Synechococcus elongatus PCC 7942 complexed with adenosine diphosphate and glucose 6-phosphate | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6-O-phosphono-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Yu, A, Xie, Y, Li, M. | | Deposit date: | 2019-07-05 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.506139 Å) | | Cite: | Photosynthetic Phosphoribulokinase Structures: Enzymatic Mechanisms and the Redox Regulation of the Calvin-Benson-Bassham Cycle.
Plant Cell, 32, 2020
|
|
4EGD
 
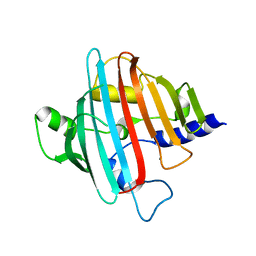 | | 1.85 Angstrom crystal structure of native hypothetical protein SAOUHSC_02783 from Staphylococcus aureus | | Descriptor: | CALCIUM ION, CHLORIDE ION, Uncharacterized protein SAOUHSC_02783 | | Authors: | Biancucci, M, Minasov, G, Halavaty, A, Filippova, E.V, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-30 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom crystal structure of native hypothetical protein SAOUHSC_02783 from Staphylococcus aureus
TO BE PUBLISHED
|
|
5DXK
 
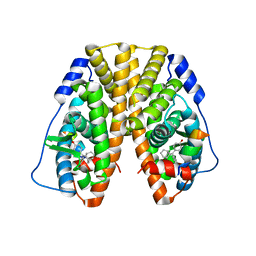 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-[(9s)-bicyclo[3.3.1]non-9-ylmethanediyl]diphenol | | Descriptor: | 4,4'-[(9s)-bicyclo[3.3.1]non-9-ylmethanediyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DWG
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Triaryl-substituted Imine Analog, 4-{(E)-(4-hydroxyphenyl)[(2-methylphenyl)imino]methyl}benzene-1,3-diol | | Descriptor: | 4-{(E)-(4-hydroxyphenyl)[(2-methylphenyl)imino]methyl}benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Wright, N.J, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-22 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
4MJM
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Short Internal Deletion of CBS Domain from Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, Inosine-5'-monophosphate dehydrogenase | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-03 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2544 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Short Internal Deletion of CBS Domain from Bacillus anthracis str. Ames
To be Published, 2013
|
|
5E3D
 
 | | Crystal structure of the human BRPF1 bromodomain in complex with SEED7 | | Descriptor: | 2,8-dithioxo-1,2,3,7,8,9-hexahydro-6H-purin-6-one, NITRATE ION, Peregrin | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2015-10-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Twenty Crystal Structures of Bromodomain and PHD Finger Containing Protein 1 (BRPF1)/Ligand Complexes Reveal Conserved Binding Motifs and Rare Interactions.
J.Med.Chem., 59, 2016
|
|
6KOC
 
 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis complexed with 3-iodo-N-oxo-2-heptyl-4-hydroxyquinoline | | Descriptor: | 2-heptyl-3-iodanyl-1-oxidanyl-quinolin-4-one, AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, ... | | Authors: | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
