8UZM
 
 | |
8UZO
 
 | |
8USI
 
 | |
5LBW
 
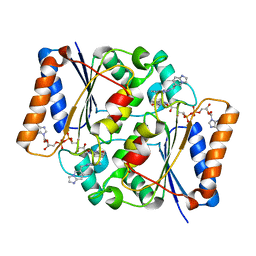 | | Structure of the human quinone reductase 2 (NQO2) in complex with volitinib | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ZINC ION, ... | | Authors: | Schneider, S, Medard, G, Kuester, B. | | Deposit date: | 2016-06-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
8UZI
 
 | |
8USL
 
 | |
8USH
 
 | |
8UQO
 
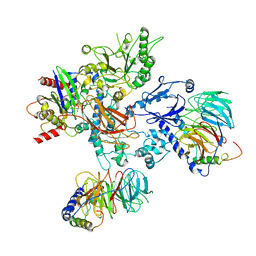 | | PLCb3-Gbg-Gaq complex on membranes | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Falzone, M.E, MacKinnon, R. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | The mechanism of G alpha q regulation of PLC beta 3 -catalyzed PIP2 hydrolysis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8USJ
 
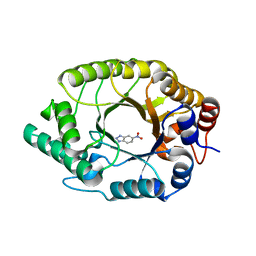 | |
8USF
 
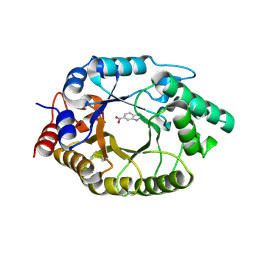 | |
8USG
 
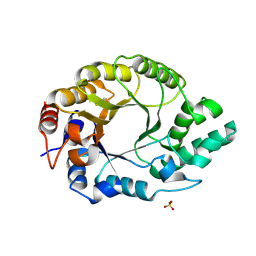 | |
6AB5
 
 | | Cryo-EM structure of T=1 Penaeus vannamei nodavirus | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Miyazaki, N, Yoshimura, M, Guan, H.H, Lin, C.C, Iwasaki, K, Chen, C.J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
8UQN
 
 | | PLCb3-Gaq complex on membranes | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Falzone, M.E, MacKinnon, R. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The mechanism of G alpha q regulation of PLC beta 3 -catalyzed PIP2 hydrolysis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5LBH
 
 | | Crystal structure of Helicobacter cinaedi CAIP | | Descriptor: | CAIP, FE (III) ION | | Authors: | Zanotti, G, Valesse, F, Codolo, G, De Bernard, M. | | Deposit date: | 2016-06-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | The Helicobacter cinaedi antigen CAIP participates in atherosclerotic inflammation by promoting the differentiation of macrophages in foam cells.
Sci Rep, 7, 2017
|
|
8UZK
 
 | |
3HPM
 
 | |
8UR3
 
 | |
8UR6
 
 | |
5LIE
 
 | |
1YCK
 
 | |
8V44
 
 | |
8VJ3
 
 | |
5LUH
 
 | | AadA E87Q in complex with ATP, calcium and streptomycin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Stern, A.L, Van der Verren, S.E, Selmer, M. | | Deposit date: | 2016-09-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural mechanism of AadA, a dual-specificity aminoglycoside adenylyltransferase fromSalmonella enterica.
J.Biol.Chem., 293, 2018
|
|
5LBZ
 
 | | Structure of the human quinone reductase 2 (NQO2) in complex with pacritinib | | Descriptor: | 11-(2-pyrrolidin-1-yl-ethoxy)-14,19-dioxa-5,7,26-triaza-tetracyclo[19.3.1.1(2,6).1(8,12)]heptacosa-1(25),2(26),3,5,8,10,12(27),16,21,23-decaene, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Schneider, S, Medard, G, Kuster, B. | | Deposit date: | 2016-06-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
5LD2
 
 | | Cryo-EM structure of RecBCD+DNA complex revealing activated nuclease domain | | Descriptor: | Fork-Hairpin DNA (70-MER), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Wilkinson, M, Chaban, Y, Wigley, D.B. | | Deposit date: | 2016-06-23 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Mechanism for nuclease regulation in RecBCD.
Elife, 5, 2016
|
|
