4CY6
 
 | | apo structure of 2-hydroxybiphenyl 3-monooxygenase HbpA | | Descriptor: | 2-HYDROXYBIPHENYL-3-MONOOXYGENASE | | Authors: | Jensen, C.N, Farrugia, J.E, Frank, A, Man, H, Hart, S, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2014-04-10 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structures of the Apo and Fad-Bound Forms of 2-Hydroxybiphenyl 3-Monooxygenase (Hbpa) Locate Activity Hotspots Identified by Using Directed Evolution.
Chembiochem, 16, 2015
|
|
2LRA
 
 | | NMR Structure of Signal Sequence Deleted (SSD) Rv0603 Protein from Mycobacterium tuberculosis without N-terminal His-tag | | Descriptor: | POSSIBLE EXPORTED PROTEIN | | Authors: | Tripathi, S, Pulavarti, S, Yadav, R, Jain, A, Pathak, P, Meher, A, Arora, A. | | Deposit date: | 2012-03-28 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Signal Sequence Deleted (SSD) Rv0603 Protein from Mycobacterium tuberculosis without N-terminal His-tag
To be Published
|
|
2LI5
 
 | | NMR structure of Atg8-Atg7C30 complex | | Descriptor: | Autophagy-related protein 8, Ubiquitin-like modifier-activating enzyme ATG7 | | Authors: | Kumeta, H, Satoo, K, Noda, N.N, Fujioka, Y, Ogura, K, Nakatogawa, H, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2011-08-23 | | Release date: | 2011-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of Atg8 activation by a homodimeric E1, Atg7.
Mol.Cell, 44, 2011
|
|
4D3K
 
 | | Structure of Bacillus subtilis nitric oxide synthase in complex with 6,6'-((5-(3-aminopropyl)-1,3-phenylene)bis(ethane-2,1-diyl))bis(4- methylpyridin-2-amine) | | Descriptor: | 6,6'-{[5-(3-aminopropyl)benzene-1,3-diyl]diethane-2,1-diyl}bis(4-methylpyridin-2-amine), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.017 Å) | | Cite: | Structure-Based Design of Bacterial Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 58, 2015
|
|
2L8N
 
 | |
4D2M
 
 | | Vaccinia Virus F1L bound to Bim BH3 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, BCL-2-LIKE PROTEIN 11, CHLORIDE ION, ... | | Authors: | Kvansakul, M, Colman, P.M. | | Deposit date: | 2014-05-12 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insight Into Bh3-Domain Binding of Vaccinia Virus Anti-Apoptotic F1L.
J.Virol., 88, 2014
|
|
6VQW
 
 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF8 | | Descriptor: | AcrF8, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2A0W
 
 | | Structure of human purine nucleoside phosphorylase H257G mutant | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Murkin, A.S, Shi, W, Schramm, V.L. | | Deposit date: | 2005-06-17 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Neighboring group participation in the transition state of human purine nucleoside phosphorylase.
Biochemistry, 46, 2007
|
|
4D3V
 
 | |
6VPH
 
 | |
6VUH
 
 | | APO PreQ1 riboswitch aptamer grown in Mn2+ | | Descriptor: | MANGANESE (II) ION, PREQ1 RIBOSWITCH | | Authors: | Jenkins, J.L, Wedekind, J.E. | | Deposit date: | 2020-02-15 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Analysis of a preQ1-I riboswitch in effector-free and bound states reveals a metabolite-programmed nucleobase-stacking spine that controls gene regulation.
Nucleic Acids Res., 48, 2020
|
|
2L6M
 
 | |
4M7E
 
 | | Structural insight into BL-induced activation of the BRI1-BAK1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1, ... | | Authors: | Chai, J, Han, Z, Sun, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.602 Å) | | Cite: | Structural insight into BL-induced activation of the BRI1-BAK1 complex
To be Published
|
|
4MH3
 
 | | Crystal structure of Bovine Mitochondrial Peroxiredoxin III | | Descriptor: | CITRIC ACID, PHOSPHATE ION, Thioredoxin-dependent peroxide reductase, ... | | Authors: | Cao, Z, McGow, D.P, Shepherd, C, Lindsay, J.G. | | Deposit date: | 2013-08-29 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Improved Catenated Structures of Bovine Peroxiredoxin III F190L Reveal Details of Ring-Ring Interactions and a Novel Conformational State.
Plos One, 10, 2015
|
|
2A83
 
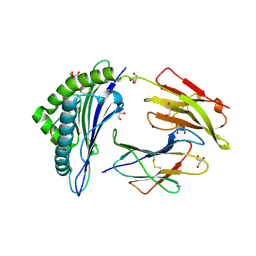 | | Crystal structure of hla-b*2705 complexed with the glucagon receptor (gr) peptide (residues 412-420) | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Ruckert, C, Fiorillo, M.T, Loll, B, Moretti, R, Biesiadka, J, Saenger, W, Ziegler, A, Sorrentino, R, Uchanska-Ziegler, B. | | Deposit date: | 2005-07-07 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational dimorphism of self-peptides and molecular mimicry in a disease-associated HLA-B27 subtype.
J.Biol.Chem., 281, 2006
|
|
4MH5
 
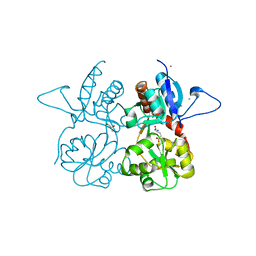 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding site and interlobe interactions of the ionotropic glutamate receptor GluK3 ligand binding domain revealed by high resolution crystal structure in complex with (S)-glutamate.
J.Struct.Biol., 176, 2011
|
|
2LE8
 
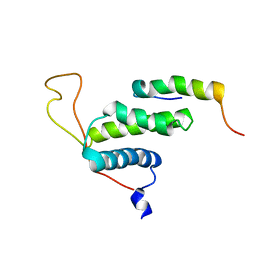 | | The protein complex for DNA replication | | Descriptor: | DNA replication factor Cdt1, DNA replication licensing factor MCM6 | | Authors: | Liu, C, Wei, Z, Zhu, G. | | Deposit date: | 2011-06-14 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the Cdt1-mediated MCM2-7 chromatin loading
Nucleic Acids Res., 40, 2012
|
|
6VXF
 
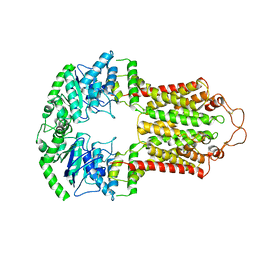 | | Structure of apo-closed ABCG2 | | Descriptor: | Broad substrate specificity ATP-binding cassette transporter ABCG2 | | Authors: | Orlando, B.J, Maofu, L. | | Deposit date: | 2020-02-21 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | ABCG2 transports anticancer drugs via a closed-to-open switch.
Nat Commun, 11, 2020
|
|
4CN8
 
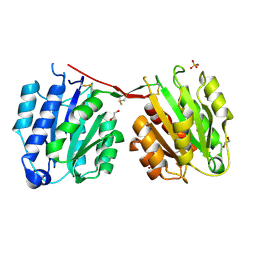 | | Structure of proximal thread matrix protein 1 (PTMP1) from the mussel byssus | | Descriptor: | 1,2-ETHANEDIOL, PROXIMAL THREAD MATRIX PROTEIN 1, SULFATE ION | | Authors: | Gertz, M, Suhre, M.H, Scheibel, T, Steegborn, C. | | Deposit date: | 2014-01-21 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and Functional Features of a Collagen-Binding Matrix Protein from the Mussel Byssus.
Nat.Commun., 5, 2014
|
|
2ABM
 
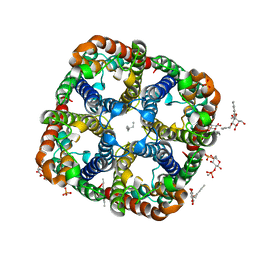 | | Crystal Structure of Aquaporin Z Tetramer Reveals both Open and Closed Water-conducting Channels | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-O-octyl-beta-D-glucopyranose, ... | | Authors: | Jiang, J, Daniels, B.V, Fu, D. | | Deposit date: | 2005-07-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of AqpZ Tetramer Reveals Two Distinct Arg-189 Conformations Associated with Water Permeation through the Narrowest Constriction of the Water-conducting Channel.
J.Biol.Chem., 281, 2006
|
|
2LA8
 
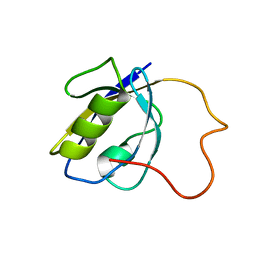 | |
4CNB
 
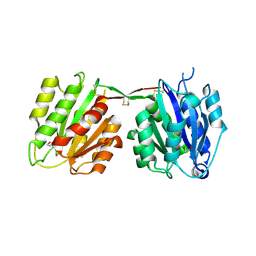 | | Structure of proximal thread matrix protein 1 (PTMP1) from the mussel byssus - Crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, PROXIMAL THREAD MATRIX PROTEIN 1, SULFATE ION | | Authors: | Gertz, M, Suhre, M.H, Scheibel, T, Steegborn, C. | | Deposit date: | 2014-01-21 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Functional Features of a Collagen-Binding Matrix Protein from the Mussel Byssus.
Nat.Commun., 5, 2014
|
|
6N46
 
 | |
4MEY
 
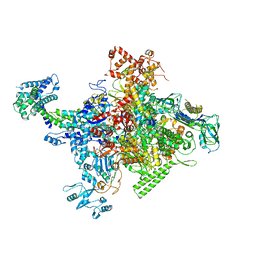 | | Crystal structure of Escherichia coli RNA polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Feng, Y, Zhang, Y, Arnold, E, Ebright, R.H. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.948 Å) | | Cite: | Transcription inhibition by the depsipeptide antibiotic salinamide A.
Elife, 3, 2014
|
|
4CS1
 
 | |
