8UQE
 
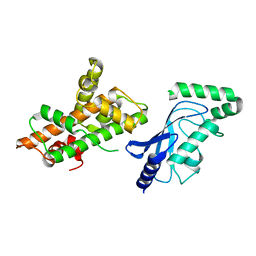 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 26-residue linker (RING not modeled in density) | | Descriptor: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.562 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQA
 
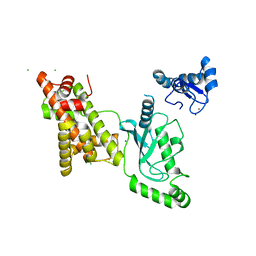 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 12-residue linker | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, SODIUM ION, ... | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQB
 
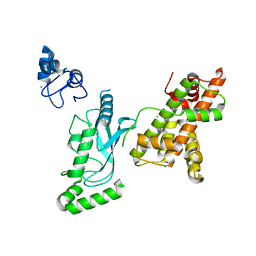 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (crystallization condition 1) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, ZINC ION | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.484 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
6SC2
 
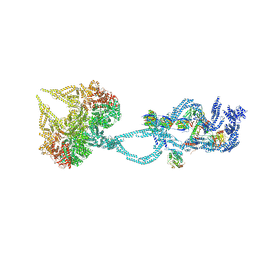 | | Structure of the dynein-2 complex; IFT-train bound model | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 2 light intermediate chain 1, ... | | Authors: | Toropova, K, Zalyte, R, Mukhopadhyay, A.G, Mladenov, M, Carter, A.P, Roberts, A.J. | | Deposit date: | 2019-07-23 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the dynein-2 complex and its assembly with intraflagellar transport trains.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8UQ9
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 4-residue linker | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, GLYCEROL, ... | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8SFN
 
 | |
8SFO
 
 | |
1BGF
 
 | |
5OKC
 
 | |
3Q3D
 
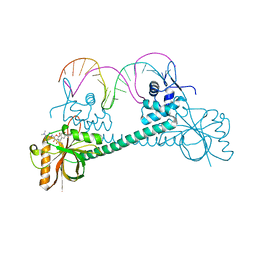 | | Crystal structure of BmrR bound to puromycin | | Descriptor: | BmrR promoter DNA, Multidrug-efflux transporter 1 regulator, PUROMYCIN | | Authors: | Bachas, S, Eginton, C, Gunio, G, Wade, H. | | Deposit date: | 2010-12-21 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural contributions to multidrug recognition in the multidrug resistance (MDR) gene regulator, BmrR.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3MXI
 
 | |
3MXM
 
 | |
2XIG
 
 | | The structure of the Helicobacter pylori ferric uptake regulator Fur reveals three functional metal binding sites | | Descriptor: | CITRIC ACID, FERRIC UPTAKE REGULATION PROTEIN, ZINC ION | | Authors: | Dian, C, Vitale, S, Leonard, G.A, Fauquant, F, Muller, C, Bahlawane, C, de Reuse, H, Michaud-Soret, I, Terradot, L. | | Deposit date: | 2010-06-29 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of the Helicobacter Pylori Ferric Uptake Regulator Fur Reveals Three Functional Metal Binding Sites.
Mol.Microbiol., 79, 2011
|
|
7ZN7
 
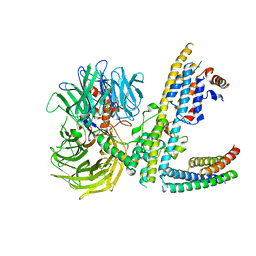 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and rat STAT2 CCD | | Descriptor: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | Authors: | Lauer, S, Spahn, C.M.T, Schwefel, D. | | Deposit date: | 2022-04-20 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
7ZNN
 
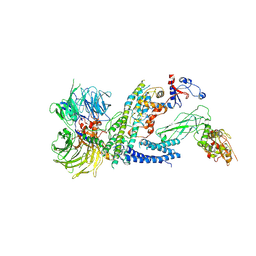 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and full-length rat STAT2 | | Descriptor: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | Authors: | Lauer, S, Spahn, C.M.T, Schwefel, D. | | Deposit date: | 2022-04-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
6C9T
 
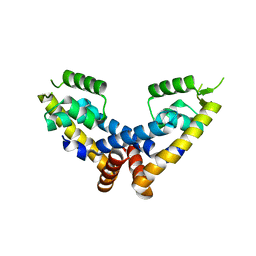 | | Transcriptional repressor, CouR | | Descriptor: | CouR | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2018-01-28 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis of transcriptional regulation by CouR, a repressor of coumarate catabolism, inRhodopseudomonas palustris.
J. Biol. Chem., 293, 2018
|
|
2JEX
 
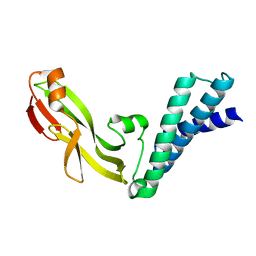 | | Transcription activator structure reveals redox control of a replication initiation reaction | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Sanders, C.M, Sizov, D, Seavers, P.R, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2007-01-24 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Transcription Activator Structure Reveals Redox Control of a Replication Initiation Reaction.
Nucleic Acids Res., 35, 2007
|
|
3NCT
 
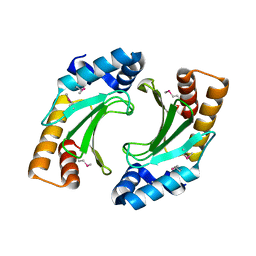 | | X-ray crystal structure of the bacterial conjugation factor PsiB, a negative regulator of reca | | Descriptor: | Protein psiB | | Authors: | Petrova, V, Satyshur, K.A, George, N.P, McCaslin, D, Cox, M.M, Keck, J.L. | | Deposit date: | 2010-06-05 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystal structure of the bacterial conjugation factor PsiB, a negative regulator of RecA.
J.Biol.Chem., 285, 2010
|
|
5CRL
 
 | |
2JEU
 
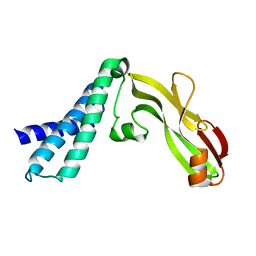 | | Transcription activator structure reveals redox control of a replication initiation reaction | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Sanders, C.M, Sizov, D, Seavers, P.R, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2007-01-23 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Transcription Activator Structure Reveals Redox Control of a Replication Initiation Reaction.
Nucleic Acids Res., 35, 2007
|
|
6C28
 
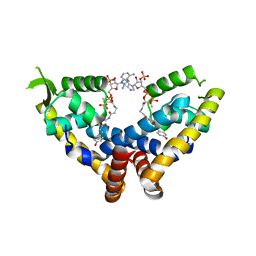 | | Transcriptional repressor, CouR, bound to p-coumaroyl-CoA | | Descriptor: | Transcriptional regulator, MarR family, p-coumaroyl-CoA | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2018-01-07 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural basis of transcriptional regulation by CouR, a repressor of coumarate catabolism, inRhodopseudomonas palustris.
J. Biol. Chem., 293, 2018
|
|
3NQU
 
 | |
7ET5
 
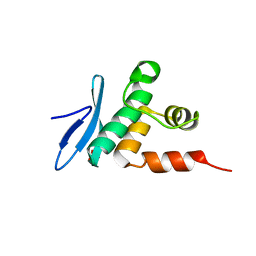 | | Crystal structure of Arabidopsis TEM1 AP2 domain | | Descriptor: | AP2/ERF and B3 domain-containing transcription repressor TEM1, SULFATE ION | | Authors: | Hu, H, Du, J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.052 Å) | | Cite: | TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4OAY
 
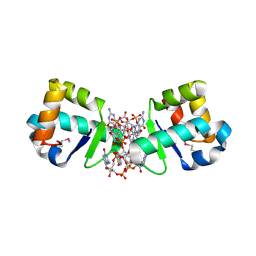 | | BldD CTD-c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein | | Authors: | Schumacher, M.A, Tschowri, N, Buttner, M, Brennan, R.G. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development.
Cell(Cambridge,Mass.), 158, 2014
|
|
3MNN
 
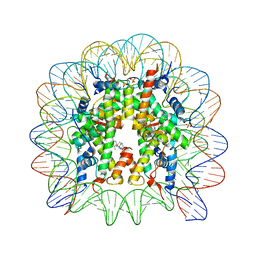 | | A Ruthenium Antitumour Agent Forms Specific Histone Protein Adducts in the Nucleosome Core | | Descriptor: | 1,3,5-triaza-7-phosphatricyclo[3.3.1.1~3,7~]decane, 1-methyl-4-(1-methylethyl)benzene, DNA (145-MER), ... | | Authors: | Ong, M.S, Davey, C.A. | | Deposit date: | 2010-04-22 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A ruthenium antimetastasis agent forms specific histone protein adducts in the nucleosome core
Chemistry, 17, 2011
|
|
