7CXC
 
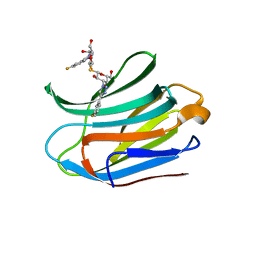 | |
8OXX
 
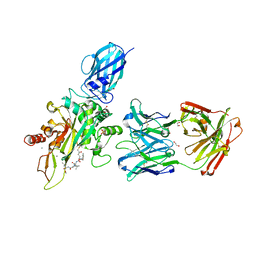 | |
6G84
 
 | | Structure of Cdc14 bound to CBK1 PxL motif | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CBK1, ... | | Authors: | Mouilleron, S, Kataria, M, Uhlmann, F. | | Deposit date: | 2018-04-07 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | A PxL motif promotes timely cell cycle substrate dephosphorylation by the Cdc14 phosphatase.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5VM6
 
 | | The hapten triclocarban bound to the single domain camelid nanobody VHH T10 | | Descriptor: | N-(4-chlorophenyl)-N'-(3,4-dichlorophenyl)urea, SODIUM ION, SULFATE ION, ... | | Authors: | Tabares-da Rosa, S, Gonzalez-Sapienza, G, Wilson, D.K. | | Deposit date: | 2017-04-26 | | Release date: | 2018-05-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and specificity of several triclocarban-binding single domain camelid antibody fragments.
J. Mol. Recognit., 32, 2019
|
|
6VBE
 
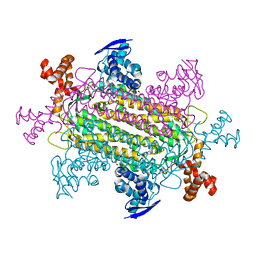 | |
8OYX
 
 | | De novo designed soluble GPCR-like fold GLF_18 | | Descriptor: | De novo designed soluble GPCR-like protein, PHOSPHATE ION | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
6N6W
 
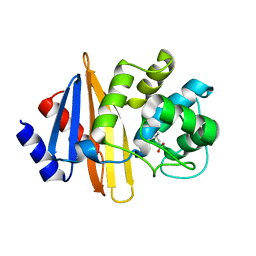 | | OXA-23 mutant F110A/M221A neutral pH form | | Descriptor: | Beta-lactamase oxa23 | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2018-11-27 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Role of the Hydrophobic Bridge in the Carbapenemase Activity of Class D beta-Lactamases.
Antimicrob. Agents Chemother., 63, 2019
|
|
5H62
 
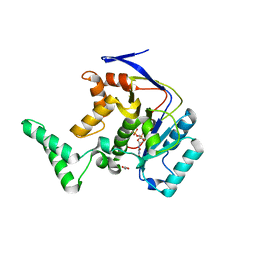 | | Structure of Transferase mutant-C23S,C199S | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, Transferase, ... | | Authors: | Park, J.B, Yoo, Y, Kim, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
5L8Y
 
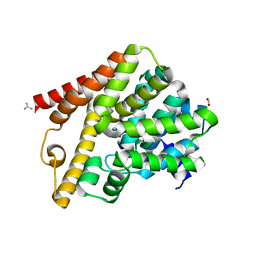 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-937 | | Descriptor: | 2-[(4-{5-[(4aR,8aS)-3-cycloheptyl-4-oxo-3,4,4a,5,8,8a-hexahydrophthalazin-1-yl]-2-methoxyphenyl}phenyl)formamido]-N-(2-hydroxyethyl)acetamide, FORMIC ACID, GLYCEROL, ... | | Authors: | Singh, A.K, Anthonyrajah, E.S, Brown, D.G. | | Deposit date: | 2016-06-08 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
7CXD
 
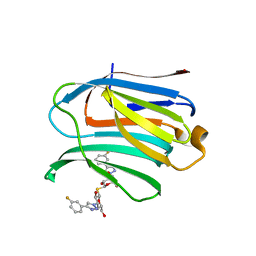 | | Xray structure of rat Galectin-3 CRD in complex with TD-139 belonging to P121 space group | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, BROMIDE ION, Galectin-3 | | Authors: | Kumar, A. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Molecular mechanism of interspecies differences in the binding affinity of TD139 to Galectin-3.
Glycobiology, 31, 2021
|
|
6PJ8
 
 | |
5HLY
 
 | |
7CWP
 
 | |
6N10
 
 | | Crystal structure of Arabidopsis thaliana mevalonate 5-diphosphate decarboxylase 1 complexed with (R)-MVAPP | | Descriptor: | (3R)-3-HYDROXY-5-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}-3-METHYLPENTANOIC ACID, Diphosphomevalonate decarboxylase MVD1, peroxisomal, ... | | Authors: | Noel, J.P, Thomas, S.T, Louie, G.V. | | Deposit date: | 2018-11-07 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate Specificity and Engineering of Mevalonate 5-Phosphate Decarboxylase.
Acs Chem.Biol., 14, 2019
|
|
6VD8
 
 | |
6FPG
 
 | | Structure of the Ustilago maydis chorismate mutase 1 in complex with a Zea mays kiwellin | | Descriptor: | CITRIC ACID, Chromosome 16, whole genome shotgun sequence, ... | | Authors: | Altegoer, F, Steinchen, W, Bange, G. | | Deposit date: | 2018-02-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A kiwellin disarms the metabolic activity of a secreted fungal virulence factor.
Nature, 565, 2019
|
|
6YGG
 
 | | NADase from Aspergillus fumigatus complexed with a substrate anologue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, AfNADase, ... | | Authors: | Stromland, O, Ziegler, M, Kallio, J.P. | | Deposit date: | 2020-03-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of fungal surface NADases predominantly present in pathogenic species.
Nat Commun, 12, 2021
|
|
5L97
 
 | |
6PJP
 
 | |
6N1A
 
 | | Crystal structure of an N-acetylgalactosamine deacetylase from F. plautii | | Descriptor: | CALCIUM ION, Carbohydrate-binding protein, GLYCEROL, ... | | Authors: | Sim, L, Rahfeld, P, Withers, S.G. | | Deposit date: | 2018-11-08 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An enzymatic pathway in the human gut microbiome that converts A to universal O type blood.
Nat Microbiol, 4, 2019
|
|
7CX6
 
 | |
8ORK
 
 | | cyclic 2,3-diphosphoglycerate synthetase from the hyperthermophilic archaeon Methanothermus fervidus | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | De Rose, S.A, Isupov, M. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural characterization of a novel cyclic 2,3-diphosphoglycerate synthetase involved in extremolyte production in the archaeon Methanothermus fervidus .
Front Microbiol, 14, 2023
|
|
6FQM
 
 | | 3.06A COMPLEX OF S.AUREUS GYRASE with imidazopyrazinone T1 AND DNA | | Descriptor: | 7-[(3~{S})-3-azanylpyrrolidin-1-yl]-5-cyclopropyl-8-fluoranyl-imidazo[1,2-a]quinoxalin-4-one, DNA (5'-D(*GP*AP*GP*AP*GP*TP*AP*T*GP*GP*CP*CP*AP*TP*AP*CP*TP*CP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Germe, T, Basque, E, Maxwell, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A new class of antibacterials, the imidazopyrazinones, reveal structural transitions involved in DNA gyrase poisoning and mechanisms of resistance.
Nucleic Acids Res., 46, 2018
|
|
5H82
 
 | | HETEROYOHIMBINE SYNTHASE THAS2 FROM CATHARANTHUS ROSEUS - APO FORM | | Descriptor: | ZINC ION, heteroyohimbine synthase THAS2 | | Authors: | Stavrinides, A, Tatsis, E.C, Caputi, L, Foureau, E, Stevenson, C.E.M, Lawson, D.M, Courdavault, V, O'Connor, S.E. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity.
Nat Commun, 7, 2016
|
|
5KM1
 
 | |
