3Q5V
 
 | |
3Q6D
 
 | | Xaa-Pro dipeptidase from Bacillus anthracis. | | Descriptor: | CALCIUM ION, GLYCEROL, Proline dipeptidase | | Authors: | Osipiuk, J, Makowska-Grzyska, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-31 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Xaa-Pro dipeptidase from Bacillus anthracis.
To be Published
|
|
3QR1
 
 | | Crystal Structure of L. pealei PLC21 | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE C-BETA (PLC-BETA) | | Authors: | Lyon, A.M, Suddala, K.C, Northup, J.K, Tesmer, J.J.G. | | Deposit date: | 2011-02-16 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | An autoinhibitory helix in the C-terminal region of phospholipase C-beta mediates Galphaq activation.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3Q60
 
 | |
3Q7Z
 
 | | CBAP-acylated BlaR1 sensor domain from Staphylococcus aureus | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2'-carboxybiphenyl-2-yl)carbonyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase regulatory protein BlaR1 | | Authors: | Borbulevych, O.Y, Mobashery, S, Baker, B.M. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Lysine Nzeta-decarboxylation switch and activation of the beta-lactam sensor domain of BlaR1 protein of methicillin-resistant Staphylococcus aureus.
J.Biol.Chem., 286, 2011
|
|
3Q6K
 
 | | Salivary protein from Lutzomyia longipalpis | | Descriptor: | 43.2 kDa salivary protein, CITRIC ACID, SEROTONIN | | Authors: | Andersen, J.F, Xu, X, Chang, B.W, Collin, N, Valenzuela, J.G, Ribeiro, J.M. | | Deposit date: | 2011-01-02 | | Release date: | 2011-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure and function of a "yellow" protein from saliva of the sand fly Lutzomyia longipalpis that confers protective immunity against Leishmania major infection.
J.Biol.Chem., 286, 2011
|
|
3Q96
 
 | | B-Raf kinase domain in complex with a tetrahydronaphthalene inhibitor | | Descriptor: | (2S)-N-[3-(2-aminopropan-2-yl)-5-(trifluoromethyl)phenyl]-7-[(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-4-yl)oxy]-1,2,3,4-tetrahydronaphthalene-2-carboxamide, Serine/threonine-protein kinase B-raf | | Authors: | Sintchak, M.D, Aertgeerts, K, Yano, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Design and optimization of potent and orally bioavailable tetrahydronaphthalene raf inhibitors.
J.Med.Chem., 54, 2011
|
|
3Q9H
 
 | | LVFFA segment from Alzheimer's Amyloid-Beta displayed on 42-membered macrocycle scaffold | | Descriptor: | 1,4-BUTANEDIOL, Cyclic pseudo-peptide LVFFA(ORN)(HAO)LK(ORN), GLYCEROL, ... | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
3Q9X
 
 | | Crystal structure of human CK2 alpha in complex with emodin at pH 6.5 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, ... | | Authors: | Battistutta, R, Ranchio, A, Papinutto, E. | | Deposit date: | 2011-01-10 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of the flexible regions of the catalytic alpha-subunit of protein kinase CK2
To be Published
|
|
3QBM
 
 | |
3QCK
 
 | |
3QCU
 
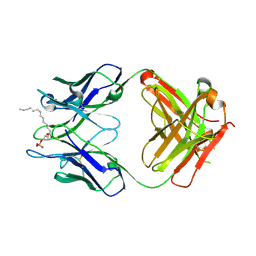 | | Crystal structure of the LT3015 antibody Fab fragment in complex with lysophosphatidic acid (14:0) | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, LT3015 antibody Fab fragment, heavy chain, ... | | Authors: | Fleming, J.K, Wojciak, J.M, Campbell, M.-A, Huxford, T. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Biochemical and structural characterization of lysophosphatidic Acid binding by a humanized monoclonal antibody.
J.Mol.Biol., 408, 2011
|
|
3Q9L
 
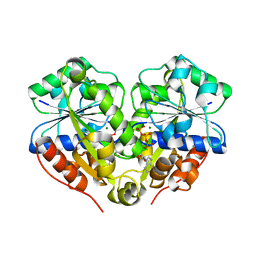 | | The structure of the dimeric E.coli MinD-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Septum site-determining protein minD | | Authors: | Wu, W, Park, K.-T, Lutkenhaus, J, Holyoak, T. | | Deposit date: | 2011-01-08 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Determination of the structure of the MinD-ATP complex reveals the orientation of MinD on the membrane and the relative location of the binding sites for MinE and MinC.
Mol.Microbiol., 79, 2011
|
|
3Q9Y
 
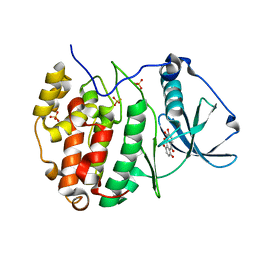 | | Crystal structure of human CK2 alpha in complex with Quinalizarin at pH 8.5 | | Descriptor: | 1,2,5,8-tetrahydroxyanthracene-9,10-dione, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Battistutta, R, Ranchio, A, Papinutto, E. | | Deposit date: | 2011-01-10 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analysis of the flexible regions of the catalytic alpha-subunit of protein kinase CK2
To be Published
|
|
3QC3
 
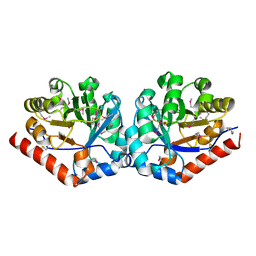 | |
3QH2
 
 | | Crystal structure of TenI from Bacillus subtilis complexed with product cThz-P | | Descriptor: | 4-methyl-5-[2-(phosphonooxy)ethyl]-1,3-thiazole-2-carboxylic acid, Regulatory protein tenI, SULFATE ION | | Authors: | Han, Y, Zhang, Y, Hazra, A, Chatterjee, A, Lai, R, Begley, T.P, Ealick, S.E. | | Deposit date: | 2011-01-25 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | A Missing Enzyme in Thiamin Thiazole Biosynthesis: Identification of TenI as a Thiazole Tautomerase.
J.Am.Chem.Soc., 133, 2011
|
|
3QCI
 
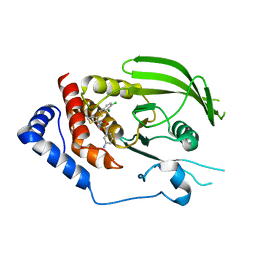 | |
3QCS
 
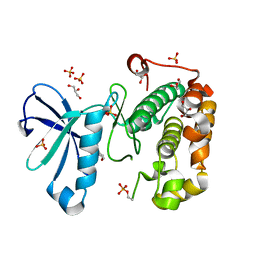 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 6-[2-Amino-6-(4-morpholinyl)-4-pyrimidinyl]-1H-indazol-3-amine | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 6-[2-amino-6-(morpholin-4-yl)pyrimidin-4-yl]-2H-indazol-3-amine, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QI7
 
 | |
3QD6
 
 | | Crystal structure of the CD40 and CD154 (CD40L) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD40 ligand, Tumor necrosis factor receptor superfamily member 5 | | Authors: | Lee, J.-O, Kim, Y.J, Song, D.H, Kim, H.M, Park, B.S. | | Deposit date: | 2011-01-18 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallographic and mutational analysis of the CD40-CD154 complex and its implications for receptor activation
J.Biol.Chem., 286, 2011
|
|
3QLR
 
 | | Candida albicans dihydrofolate reductase complexed with NADPH and 6-methyl-5-[(3R)-3-(3,4,5-trimethoxyphenyl)pent-1-yn-1-yl]pyrimidine-2,4-diamine (UCP112A) | | Descriptor: | 6-methyl-5-[(3R)-3-(3,4,5-trimethoxyphenyl)pent-1-yn-1-yl]pyrimidine-2,4-diamine, GLYCEROL, GLYCINE, ... | | Authors: | Paulsen, J.L, Bendel, S.D, Anderson, A.C. | | Deposit date: | 2011-02-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Crystal Structures of Candida albicans Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal the Flexibility of Active Site Loop Residues Critical for Ligand Potency and Selectivity.
Chem.Biol.Drug Des., 78, 2011
|
|
3QGE
 
 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid and (2R)-4-(2,6-dimethoxypyrimidin-4-yl)-N-(4-methoxybenzyl)-1-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazine-2-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, (2R)-4-(2,6-dimethoxypyrimidin-4-yl)-N-(4-methoxybenzyl)-1-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazine-2-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Investigation of the mode of binding of a novel series of N-benzyl-4-heteroaryl-1-(phenylsulfonyl)piperazine-2-carboxamides to the hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3QMF
 
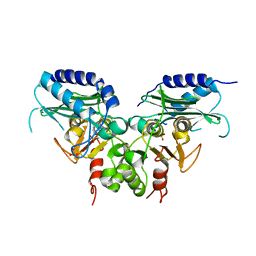 | | Crystal strucuture of an inositol monophosphatase family protein (SAS2203) from Staphylococcus aureus MSSA476 | | Descriptor: | Inositol monophosphatase family protein, SULFATE ION | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2011-02-04 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Staphylococcal dual specific inositol monophosphatase/NADP(H) phosphatase (SAS2203) delineates the molecular basis of substrate specificity
Biochimie, 94, 2012
|
|
3Q6J
 
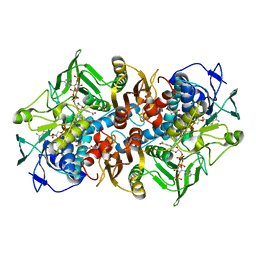 | | Structural basis for carbon dioxide binding by 2-ketopropyl coenzyme M Oxidoreductase/Carboxylase | | Descriptor: | (2-[2-KETOPROPYLTHIO]ETHANESULFONATE, 1-THIOETHANESULFONIC ACID, 2-oxopropyl-CoM reductase, ... | | Authors: | Pandey, A.S, Mulder, D.W, Ensign, S.A, Peters, J.W. | | Deposit date: | 2011-01-01 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for carbon dioxide binding by 2-ketopropyl coenzyme M oxidoreductase/carboxylase.
Febs Lett., 585, 2011
|
|
3QKB
 
 | |
