6S68
 
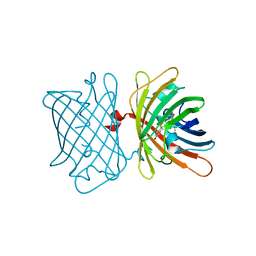 | | Structure of the Fluorescent Protein AausFP2 from Aequorea cf. australis at pH 7.6 | | Descriptor: | Aequorea cf. australis fluorescent protein 2 (AausFP2) | | Authors: | Depernet, H, Gotthard, G, Lambert, G.G, Shaner, N.C, Royant, A. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Aequorea's secrets revealed: New fluorescent proteins with unique properties for bioimaging and biosensing.
Plos Biol., 18, 2020
|
|
6S67
 
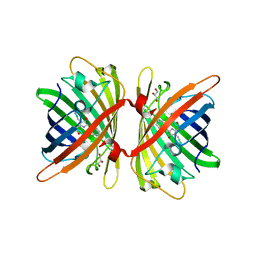 | | Structure of the Fluorescent Protein AausFP1 from Aequorea cf. australis at pH 7.0 | | Descriptor: | Aequorea cf. australis fluorescent protein 1 (AausFP1), GLYCEROL | | Authors: | Depernet, H, Gotthard, G, Lambert, G.G, Shaner, N.C, Royant, A. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Aequorea's secrets revealed: New fluorescent proteins with unique properties for bioimaging and biosensing.
Plos Biol., 18, 2020
|
|
6THT
 
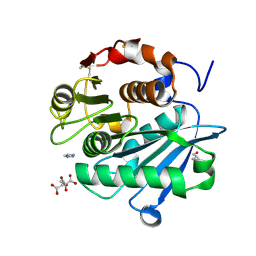 | |
6THS
 
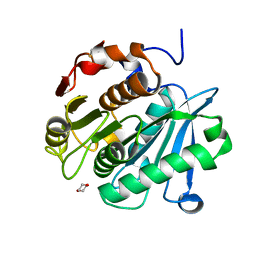 | |
6YKD
 
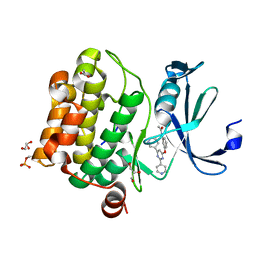 | | Human Pim-1 kinase in complex with an inhibitor identified by virtual screening | | Descriptor: | ACETATE ION, GLYCEROL, Serine/threonine-protein kinase pim-1, ... | | Authors: | Schneider, P, Welin, M, Svensson, B, Walse, B, Schneider, G. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Virtual Screening and Design with Machine Intelligence Applied to Pim-1 Kinase Inhibitors.
Mol Inform, 39, 2020
|
|
5K0X
 
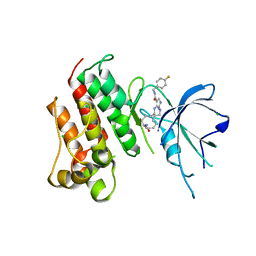 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2541 | | Descriptor: | (7S)-7-amino-N-[(4-fluorophenyl)methyl]-8-oxo-2,9,16,18,21-pentaazabicyclo[15.3.1]henicosa-1(21),17,19-triene-20-carboxamide, CHLORIDE ION, Tyrosine-protein kinase Mer | | Authors: | McIver, A.L, Zhang, W, Liu, Q, Jiang, X, Stashko, M.A, Nichols, J, Miley, M.J, Norris-Drouin, J, Machius, M, DeRyckere, D, Wood, E, Graham, D.K, Earp, H.S, Kireev, D, Frye, S.V, Wang, X. | | Deposit date: | 2016-05-17 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Discovery of Macrocyclic Pyrimidines as MerTK-Specific Inhibitors.
ChemMedChem, 12, 2017
|
|
5K2E
 
 | | Structure of NNQQNY from yeast prion Sup35 with zinc acetate determined by MicroED | | Descriptor: | ACETIC ACID, Eukaryotic peptide chain release factor GTP-binding subunit, ZINC ION | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7NDZ
 
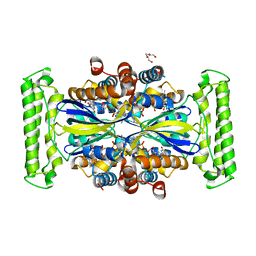 | | ThyX reconstituted with N5-carbinolamine flavin | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent thymidylate synthase, ... | | Authors: | Bou-Nader, C, Pecqueur, L, Hamdane, D. | | Deposit date: | 2021-02-02 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An enzymatic activation of formaldehyde for nucleotide methylation.
Nat Commun, 12, 2021
|
|
7NDW
 
 | | ThyX-FADH2 soaked with 20 mM Formaldehyde | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent thymidylate synthase, ... | | Authors: | Bou-Nader, C, Pecqueur, L, Hamdane, D. | | Deposit date: | 2021-02-02 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An enzymatic activation of formaldehyde for nucleotide methylation.
Nat Commun, 12, 2021
|
|
7RDR
 
 | |
4E7A
 
 | |
4GD0
 
 | | tRNA-guanine transglycosylase Y106F, C158V mutant | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Schmidt, I, Tidten-Luksch, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2012-07-31 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
4E76
 
 | |
4E78
 
 | |
4H7Z
 
 | | tRNA-guanine transglycosylase Y106F, C158V mutant in complex with guanine | | Descriptor: | GLYCEROL, GUANINE, Queuine tRNA-ribosyltransferase, ... | | Authors: | Biela, I, Tidten-Luksch, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
4H6E
 
 | | tRNA-guanine transglycosylase Y106F, C158V, V233G mutant apo structure | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Biela, I, Tidten-Luksch, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2012-09-19 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
4HSH
 
 | | tRNA-guanine transglycosylase Y106F, V233G mutant in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Biela, I, Tidten-Luksch, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2012-10-30 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
4HVX
 
 | | tRNA-guanine transglycosylase Y106F, C158V mutant in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Biela, I, Tidten-Luksch, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2012-11-07 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
4HQV
 
 | | tRNA-guanine transglycosylase Y106F, C158V, V233G mutant in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Biela, I, Tidten-Luksch, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2012-10-26 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
3P96
 
 | |
3PM6
 
 | |
3QBH
 
 | | Structure based design, synthesis and SAR of cyclic hydroxyethylamine (HEA) BACE-1 inhibitors | | Descriptor: | (4S)-4-(2-hydroxy-5-{[(3S,4S,5R)-4-hydroxy-1,1-dioxido-5-{[3-(propan-2-yl)benzyl]amino}tetrahydro-2H-thiopyran-3-yl]methyl}benzyl)-3-propyl-1,3-oxazolidin-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M. | | Deposit date: | 2011-01-13 | | Release date: | 2011-03-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure based design, synthesis and SAR of cyclic hydroxyethylamine (HEA) BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3OH0
 
 | | Protein structure of USP from L. major bound to URIDINE-5'-TRIPHOSPHATE | | Descriptor: | GLYCEROL, UDP-sugar pyrophosphorylase, URIDINE 5'-TRIPHOSPHATE | | Authors: | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F, Ficner, R. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
3OGZ
 
 | | Protein structure of USP from L. major in Apo-form | | Descriptor: | GLYCEROL, UDP-sugar pyrophosphorylase | | Authors: | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F, Ficner, R. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
3STW
 
 | |
