7DXA
 
 | | PSII intermediate Psb28-RC47 | | Descriptor: | (1S)-2-(ALPHA-L-ALLOPYRANOSYLOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYL PALMITATE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 5-[(2E,6E,10E,14E,18E,22E)-3,7,11,15,19,23,27-HEPTAMETHYLOCTACOSA-2,6,10,14,18,22,26-HEPTAENYL]-2,3-DIMETHYLBENZO-1,4-QUINONE, ... | | Authors: | Sui, S.F, Shen, J.R, Han, G.Y, Xiao, Y.N, Huang, G.Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-06-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural insights into cyanobacterial photosystem II intermediates associated with Psb28 and Tsl0063.
Nat.Plants, 7, 2021
|
|
7E14
 
 | | Compound2_GLP-1R_OWL833_Gs complex structure | | Descriptor: | 3-[(1S,2S)-1-(5-[(4S)-2,2-dimethyloxan-4-yl]-2-{(4S)-2-(4-fluoro-3,5-dimethylphenyl)-3-[3-(4-fluoro-1-methyl-1H-indazol-5-yl)-2-oxo-2,3-dihydro-1H-imidazol-1-yl]-4-methyl-2,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carbonyl}-1H-indol-1-yl)-2-methylcyclopropyl]-1,2,4-oxadiazol-5(4H)-one, CHOLESTEROL, G protein, ... | | Authors: | Cong, Z.T, Chen, L.N, Ma, H.L, Yang, D.H, Xu, H.E, Zhang, Y, Wang, M.W. | | Deposit date: | 2021-01-30 | | Release date: | 2021-07-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
7EDA
 
 | | Structure of monomeric photosystem II | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Yu, H, Hamaguchi, T, Nakajima, Y, Kato, K, kawakami, K, Akita, F, Yonekura, K, Shen, J.R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Cryo-EM structure of monomeric photosystem II at 2.78 angstrom resolution reveals factors important for the formation of dimer.
Biochim Biophys Acta Bioenerg, 1862, 2021
|
|
7EIN
 
 | | SARS-CoV-2 main proteinase complex with microbial metabolite leupeptin | | Descriptor: | 3C-like proteinase, leupeptin | | Authors: | Fu, L.F, Feng, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-03-31 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Microbial Metabolite Leupeptin in the Treatment of COVID-19 by Traditional Chinese Medicine Herbs.
Mbio, 12, 2021
|
|
7DUQ
 
 | | Cryo-EM structure of the compound 2 and GLP-1-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1, Glucagon-like peptide 1 receptor, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
7E8P
 
 | | Crystal structure of a Flavin-dependent Monooxygenase HadA wild type complexed with reduced FAD and 4-nitrophenol | | Descriptor: | Chlorophenol monooxygenase, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, P-NITROPHENOL | | Authors: | Pimviriyakul, P, Jaruwat, A, Chitnumsub, P, Chaiyen, P. | | Deposit date: | 2021-03-02 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into a flavin-dependent dehalogenase HadA explain catalysis and substrate inhibition via quadruple pi-stacking.
J.Biol.Chem., 297, 2021
|
|
7E0P
 
 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with 4-(((6-Bromo-1H-indazol-4-yl)amino)methyl)phenol (2) | | Descriptor: | 4-[[(6-bromanyl-1~{H}-indazol-4-yl)amino]methyl]phenol, ACETIC ACID, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.635 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
7E0T
 
 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with (1R,2S)-2-(((5-bromo-1H-indazol-4-yl)amino)methyl)Cyclohexan-1-ol (36) | | Descriptor: | (1~{R},2~{S})-2-[[(5-bromanyl-1~{H}-indazol-4-yl)amino]methyl]cyclohexan-1-ol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
2MCM
 
 | | MACROMOMYCIN | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Van Roey, P. | | Deposit date: | 1991-05-08 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure analysis of auromomycin apoprotein (macromomycin) shows importance of protein side chains to chromophore binding selectivity.
Proc.Natl.Acad.Sci.Usa, 86, 1989
|
|
7E0O
 
 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with 6-Bromo-1H-indazol-4-amine (1) | | Descriptor: | 6-bromanyl-1~{H}-indazol-4-amine, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.337 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
7E0S
 
 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with (1R,2S)-2-(((6-Bromo-1H-indazol-4-yl)amino)methyl)cyclohexan-1-ol (23) | | Descriptor: | (1~{R},2~{S})-2-[[(6-bromanyl-1~{H}-indazol-4-yl)amino]methyl]cyclohexan-1-ol, ACETIC ACID, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.712 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
7E0U
 
 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with 6-Bromo-N-(((1S,2S)-2-chlorocyclohexyl)methyl)-1H-indazol-4-amine (39) | | Descriptor: | 6-bromanyl-~{N}-[[(1~{S},2~{S})-2-chloranylcyclohexyl]methyl]-1~{H}-indazol-4-amine, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.278 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
7E0Q
 
 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with (1S,2R)-2-(((6-Bromo-1H-indazol-4-yl)amino)methyl)cyclohexan-1-ol (22) | | Descriptor: | (1~{S},2~{R})-2-[[(6-bromanyl-1~{H}-indazol-4-yl)amino]methyl]cyclohexan-1-ol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.462 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
4DX5
 
 | | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, Acriflavine resistance protein B, DARPIN, ... | | Authors: | Eicher, T, Cha, H, Seeger, M.A, Brandstaetter, L, El-Delik, J, Bohnert, J.A, Kern, W.V, Verrey, F, Gruetter, M.G, Diederichs, K, Pos, K.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7DUR
 
 | | Cryo-EM structure of the compound 2-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
7EGN
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | H-Bonding Networks Dictate the Molecular Mechanism of H2O2 Activation by P450
Acs Catalysis, 11, 2021
|
|
2J0D
 
 | |
5XGG
 
 | |
7E56
 
 | |
7DVE
 
 | | Crystal structure of FAD-dependent C-glycoside oxidase | | Descriptor: | 6'''-hydroxyparomomycin C oxidase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Senda, M, Watanabe, S, Kumano, T, Kobayashi, M, Senda, T. | | Deposit date: | 2021-01-13 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | FAD-dependent C -glycoside-metabolizing enzymes in microorganisms: Screening, characterization, and crystal structure analysis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EIM
 
 | |
7EJT
 
 | |
5XV6
 
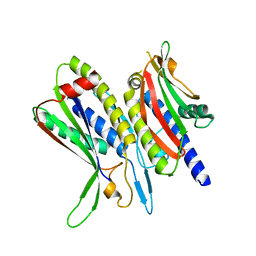 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13 | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
7EJP
 
 | |
7EKX
 
 | |
