8G2U
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:control-apo-70S at 900ms | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
1FXT
 
 | | STRUCTURE OF A CONJUGATING ENZYME-UBIQUITIN THIOLESTER COMPLEX | | Descriptor: | UBIQUITIN, UBIQUITIN-CONJUGATING ENZYME E2-24 KDA | | Authors: | Hamilton, K.S, Shaw, G.S, Williams, R.S, Huzil, J.T, McKenna, S, Ptak, C, Glover, M, Ellison, M.J. | | Deposit date: | 2000-09-26 | | Release date: | 2001-10-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail.
Structure, 9, 2001
|
|
5MWH
 
 | | Crystal structure of the human BRPF1 bromodomain in complex with BZ089 | | Descriptor: | NITRATE ION, Peregrin, ~{N}-[1,4-dimethyl-7-morpholin-4-yl-2,3-bis(oxidanylidene)quinoxalin-6-yl]-4-(2-methylpropyl)benzenesulfonamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-01-18 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-based discovery of selective BRPF1 bromodomain inhibitors.
Eur J Med Chem, 155, 2018
|
|
5MWZ
 
 | |
3QH5
 
 | | Structure of Thermolysin in complex with N-Carbobenzyloxy-L-aspartic acid and L-Phenylalanine Methyl Ester | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, N-[(benzyloxy)carbonyl]-L-aspartic acid, ... | | Authors: | Birrane, G, Bhyravbhatla, B, Navia, M. | | Deposit date: | 2011-01-25 | | Release date: | 2012-01-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis of Aspartame by Thermolysin: An X-ray Structural Study.
ACS MED.CHEM.LETT., 5, 2014
|
|
3QGO
 
 | | Structure of Thermolysin in complex with L-Phenylalanine methylester | | Descriptor: | ACETATE ION, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Birrane, G, Bhyravbhatla, B, Navia, M. | | Deposit date: | 2011-01-24 | | Release date: | 2012-01-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Synthesis of Aspartame by Thermolysin: An X-ray Structural Study.
ACS MED.CHEM.LETT., 5, 2014
|
|
3QH1
 
 | | Structure of Thermolysin in complex with N-benzyloxycarbonyl-L-aspartic acid | | Descriptor: | CALCIUM ION, N-[(benzyloxy)carbonyl]-L-aspartic acid, Thermolysin, ... | | Authors: | Birrane, G, Bhyravbhatla, B, Navia, M. | | Deposit date: | 2011-01-25 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Synthesis of Aspartame by Thermolysin: An X-ray Structural Study.
ACS MED.CHEM.LETT., 5, 2014
|
|
4LQM
 
 | | EGFR L858R in complex with PD168393 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N-[4-(3-BROMO-PHENYLAMINO)-QUINAZOLIN-6-YL]-ACRYLAMIDE | | Authors: | Yun, C.H, Eck, M.J. | | Deposit date: | 2013-07-19 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural, Biochemical, and Clinical Characterization of Epidermal Growth Factor Receptor (EGFR) Exon 20 Insertion Mutations in Lung Cancer.
Sci Transl Med, 5, 2013
|
|
6GXM
 
 | | Cryo-EM structure of an E. coli 70S ribosome in complex with RF3-GDPCP, RF1(GAQ) and Pint-tRNA (State II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Graf, M, Huter, P, Maracci, C, Peterek, M, Rodnina, M.V, Wilson, D.N. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Visualization of translation termination intermediates trapped by the Apidaecin 137 peptide during RF3-mediated recycling of RF1.
Nat Commun, 9, 2018
|
|
4LRM
 
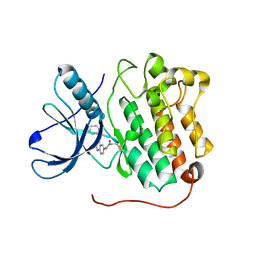 | | EGFR D770_N771insNPG in complex with PD168393 | | Descriptor: | Epidermal growth factor receptor, N-{4-[(3-bromophenyl)amino]quinazolin-6-yl}propanamide | | Authors: | Yun, C.H, Eck, M.J. | | Deposit date: | 2013-07-20 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.526 Å) | | Cite: | Structural, Biochemical, and Clinical Characterization of Epidermal Growth Factor Receptor (EGFR) Exon 20 Insertion Mutations in Lung Cancer.
Sci Transl Med, 5, 2013
|
|
6AOC
 
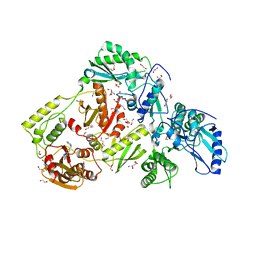 | | Crystal Structure of an N-Hydroxythienopyrimidine-2,4-dione RNase H Active Site Inhibitor with Multiple Binding Modes to HIV Reverse Transcriptase | | Descriptor: | 1,2-ETHANEDIOL, 6-benzyl-3-hydroxythieno[2,3-d]pyrimidine-2,4(1H,3H)-dione, MANGANESE (II) ION, ... | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-08-15 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, synthesis and biological evaluations of N-Hydroxy thienopyrimidine-2,4-diones as inhibitors of HIV reverse transcriptase-associated RNase H.
Eur J Med Chem, 141, 2017
|
|
6GXN
 
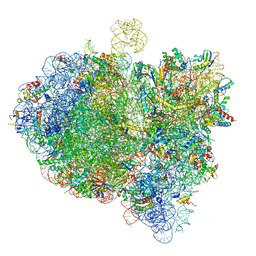 | | Cryo-EM structure of an E. coli 70S ribosome in complex with RF3-GDPCP, RF1(GAQ) and Pint-tRNA (State III) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Graf, M, Huter, P, Maracci, C, Peterek, M, Rodnina, M.V, Wilson, D.N. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Visualization of translation termination intermediates trapped by the Apidaecin 137 peptide during RF3-mediated recycling of RF1.
Nat Commun, 9, 2018
|
|
7W1M
 
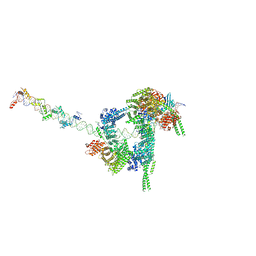 | | Cryo-EM structure of human cohesin-CTCF-DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cohesin subunit SA-1, ... | | Authors: | Shi, Z.B, Bai, X.C, Yu, H. | | Deposit date: | 2021-11-19 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | CTCF and R-loops are boundaries of cohesin-mediated DNA looping.
Mol.Cell, 83, 2023
|
|
6W2S
 
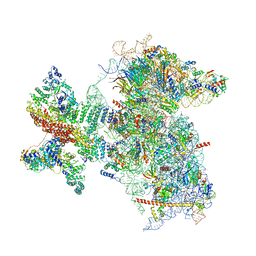 | | Structure of the Cricket Paralysis Virus 5-UTR IRES (CrPV 5-UTR-IRES) bound to the small ribosomal subunit in the open state (Class 1) | | Descriptor: | 18S rRNA, CrPV 5'-UTR IRES, Eukaryotic translation initiation factor 3 subunit A, ... | | Authors: | Neupane, R, Pisareva, V, Rodriguez, C.F, Pisarev, A, Fernandez, I.S. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | A complex IRES at the 5'-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate.
Elife, 9, 2020
|
|
5IWL
 
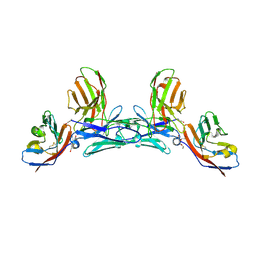 | | CD47-diabody complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5F9 diabody, Leukocyte surface antigen CD47, ... | | Authors: | Di, W, Jude, K.M, Garcia, K.C. | | Deposit date: | 2016-03-22 | | Release date: | 2016-06-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CD47-blocking immunotherapies stimulate macrophage-mediated destruction of small-cell lung cancer.
J.Clin.Invest., 126, 2016
|
|
6W2T
 
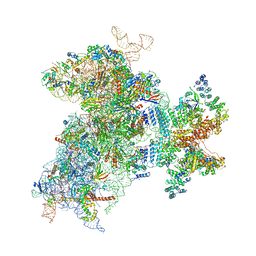 | | Structure of the Cricket Paralysis Virus 5-UTR IRES (CrPV 5-UTR-IRES) bound to the small ribosomal subunit in the closed state (Class 2) | | Descriptor: | 18S rRNA, CrPV 5'-UTR IRES, Eukaryotic translation initiation factor 3 subunit A, ... | | Authors: | Neupane, R, Pisareva, V, Rodriguez, C.F, Pisarev, A, Fernandez, I.S. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | A complex IRES at the 5'-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate.
Elife, 9, 2020
|
|
7PEQ
 
 | | Model of the outer rings of the human nuclear pore complex | | Descriptor: | Nuclear pore complex protein Nup107, Nuclear pore complex protein Nup133, Nuclear pore complex protein Nup160, ... | | Authors: | Schuller, A.P, Wojtynek, M, Mankus, D, Tatli, M, Kronenberg-Tenga, R, Regmi, S.G, Dasso, M, Weis, K, Medalia, O, Schwartz, T.U. | | Deposit date: | 2021-08-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The cellular environment shapes the nuclear pore complex architecture.
Nature, 598, 2021
|
|
7QVP
 
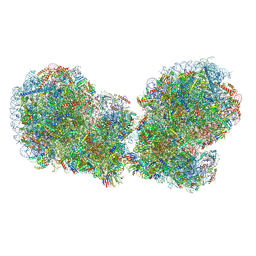 | | Human collided disome (di-ribosome) stalled on XBP1 mRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Denk, T.G, Tesina, P, Beckmann, R. | | Deposit date: | 2022-01-22 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A distinct mammalian disome collision interface harbors K63-linked polyubiquitination of uS10 to trigger hRQT-mediated subunit dissociation.
Nat Commun, 13, 2022
|
|
5M3Y
 
 | | Crystal structure of human glycosylated angiotensinogen | | Descriptor: | Angiotensinogen, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yan, Y, Read, R.J. | | Deposit date: | 2016-10-17 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the specificity of renin-mediated angiotensinogen cleavage.
J. Biol. Chem., 294, 2019
|
|
7LS5
 
 | | Cryo-EM structure of the Pre3-1 20S proteasome core particle | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Schnell, H.M, Walsh Jr, R.M, Rawson, S, Hanna, J.W. | | Deposit date: | 2021-02-17 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structures of chaperone-associated assembly intermediates reveal coordinated mechanisms of proteasome biogenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6RBF
 
 | | Mucin 2 D3 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Javitt, G, Fass, D. | | Deposit date: | 2019-04-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Intestinal Gel-Forming Mucins Polymerize by Disulfide-Mediated Dimerization of D3 Domains.
J.Mol.Biol., 431, 2019
|
|
8D3P
 
 | | Type I-C Cas4-Cas1-Cas2 complex bound to half-site integration intermediate (HSI) | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endonuclease Cas2, CRISPR-associated exonuclease Cas4, ... | | Authors: | Dhingra, Y, Suresh, S.K, Juneja, P, Sashital, D.G. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | PAM binding ensures orientational integration during Cas4-Cas1-Cas2-mediated CRISPR adaptation.
Mol.Cell, 82, 2022
|
|
8UIP
 
 | |
1B4Q
 
 | | Solution structure of human thioltransferase complex with glutathione | | Descriptor: | GLUTATHIONE, PROTEIN (HUMAN THIOLTRANSFERASE) | | Authors: | Yang, Y, Jao, S.C, Nanduri, S, Starke, D.W, Mieyal, J.J, Qin, J. | | Deposit date: | 1998-12-25 | | Release date: | 1999-12-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Reactivity of the human thioltransferase (glutaredoxin) C7S, C25S, C78S, C82S mutant and NMR solution structure of its glutathionyl mixed disulfide intermediate reflect catalytic specificity.
Biochemistry, 37, 1998
|
|
5GWK
 
 | | Human topoisomerase IIalpha in complex with DNA and etoposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(P*AP*GP*CP*CP*GP*AP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*GP*CP*TP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Wang, Y.R, Wu, C.C, Chan, N.L. | | Deposit date: | 2016-09-12 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.152 Å) | | Cite: | Producing irreversible topoisomerase II-mediated DNA breaks by site-specific Pt(II)-methionine coordination chemistry
Nucleic Acids Res., 45, 2017
|
|
